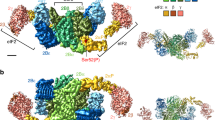
Abstract
During environmental stress, organisms limit protein synthesis by storing inactive ribosomes that are rapidly reactivated when conditions improve. Here we present structural and biochemical data showing that protein Y, an Escherichia coli stress protein, fills the tRNA- and mRNA-binding channel of the small ribosomal subunit to stabilize intact ribosomes. Protein Y inhibits translation initiation during cold shock but not at normal temperatures. Furthermore, protein Y competes with conserved translation initiation factors that, in bacteria, are required for ribosomal subunit dissociation. The mechanism used by protein Y to reduce translation initiation during stress and quickly release ribosomes for renewed translation initiation may therefore occur widely in nature.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
20 March 2007
Revised PDB code
Notes
*NOTE: In the version of this article initially published, the PDB codes are incorrect. The correct codes are 1VOQ, 1VOS, 1VOV, 1VOX, 1VOZ for the 30S and 1VOR, 1VOU, 1VOW, 1VOY and 1VP0 for the 50S. The error has been corrected in the HTML and PDF versions of the article.
References
Agafonov, D.E., Kolb, V.A. & Spirin, A.S. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2, 399–402 (2001).
Maki, Y., Yoshida, H. & Wada, A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli . Genes Cells 5, 965–974 (2000).
Ye, K., Serganov, A., Hu, W., Garber, M. & Patel, D.J. Ribosome-associated factor Y adopts a fold resembling a double-stranded RNA binding domain scaffold. Eur. J. Biochem. 269, 5182–5191 (2002).
Rak, A., Kalinin, A., Shcherbakov, D. & Bayer, P. Solution structure of the ribosome-associated cold shock response protein Yfia of Escherichia coli . Biochem. Biophys. Res. Commun. 299, 710–714 (2002).
Agafonov, D.E., Kolb, V.A., Nazimov, I.V. & Spirin, A.S. A protein residing at the subunit interface of the bacterial ribosome. Proc. Natl. Acad. Sci. USA 96, 12345–12349 (1999).
Agafonov, D.E. & Spirin, A.S. The ribosome-associated inhibitor A reduces translation errors. Biochem. Biophys. Res. Commun. 320, 354–358 (2004).
VanBogelen, R.A. & Neidhardt, F.C. Ribosomes as sensors of heat and cold shock in Escherichia coli . Proc. Natl. Acad. Sci. USA 87, 5589–5593 (1990).
Cashel, M., Gentry, D.R., Hernandez, V.J. & Vinella, D. The stringent response. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology Vol. 2 (eds. Neidhardt, F.C. et al.) 1458–1496 (ASM Press, Washington, DC, 1996).
Ashe, M.P., De Long, S.K. & Sachs, A.B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11, 833–848 (2000).
Vila-Sanjurjo, A. et al. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli . Proc. Natl. Acad. Sci. USA 100, 8682–8687 (2003).
Yusupov, M.M. et al. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 (2001).
Maivali, U. & Remme, J. Definition of bases in 23S rRNA essential for ribosomal subunit association. RNA 10, 600–604 (2004).
Yusupova, G.Z., Yusupov, M.M., Cate, J.H. & Noller, H.F. The path of messenger RNA through the ribosome. Cell 106, 233–241 (2001).
Prince, J.B., Taylor, B.H., Thurlow, D.L., Ofengand, J. & Zimmermann, R.A. Covalent crosslinking of tRNA1Val to 16S RNA at the ribosomal P site: identification of crosslinked residues. Proc. Natl. Acad. Sci. USA 79, 5450–5454 (1982).
Moazed, D. & Noller, H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148 (1989).
von Ahsen, U. & Noller, H.F. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science 267, 234–237 (1995).
Vila-Sanjurjo, A. & Dahlberg, A.E. Mutational analysis of the conserved bases C1402 and A1500 in the center of the decoding domain of Escherichia coli 16 S rRNA reveals an important tertiary interaction. J. Mol. Biol. 308, 457–463 (2001).
Wimberly, B.T. et al. Structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000).
Jacob, W.F., Santer, M. & Dahlberg, A.E. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl. Acad. Sci. USA 84, 4757–4761 (1987).
Gualerzi, C.O., Giuliodori, A.M. & Pon, C.L. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331, 527–539 (2003).
Brock, S., Szkaradkiewicz, K. & Sprinzl, M. Initiation factors of protein biosynthesis in bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol. 29, 409–417 (1998).
McCutcheon, J.P. et al. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. USA 96, 4301–4306 (1999).
Dallas, A. & Noller, H.F. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8, 855–864 (2001).
Carter, A.P. et al. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291, 498–501 (2001).
Giuliodori, A.M., Brandi, A., Gualerzi, C.O. & Pon, C.L. Preferential translation of cold-shock mRNAs during cold adaptation. RNA 10, 265–276 (2004).
Thieringer, H.A., Jones, P.G. & Inouye, M. Cold shock and adaptation. Bioessays 20, 49–57 (1998).
Uchida, T., Abe, M., Matsuo, K. & Yoneda, M. Amounts of free 70S ribosomes and ribosomal subunits found in Escherichia coli at various temperatures. Biochem. Biophys. Res. Commun. 41, 1048–1054 (1970).
Broeze, R.J., Solomon, C.J. & Pope, D.H. Effects of low temperature on in vivo and in vitro protein synthesis in Escherichia coli and Pseudomonas fluorescens . J. Bacteriol. 134, 861–874 (1978).
Barbieri, M., Vittone, A. & Maraldi, N.M. Cell stress and ribosome crystallization. J. Submicrosc. Cytol. Pathol. 27, 199–207 (1995).
Ranu, R.S., Levin, D.H., Delaunay, J., Ernst, V. & London, I.M. Regulation of protein synthesis in rabbit reticulocyte lysates: characteristics of inhibition of protein synthesis by a translational inhibitor from heme-deficient lysates and its relationship to the initiation factor which binds Met-tRNAf. Proc. Natl. Acad. Sci. USA 73, 2720–2724 (1976).
Cooper, H.L., Berger, S.L. & Braverman, R. Free ribosomes in physiologically nondividing cells. Human peripheral lymphocytes. J. Biol. Chem. 251, 4891–4900 (1976).
Kyrpides, N.C. & Woese, C.R. Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. USA 95, 224–228 (1998).
Lomakin, I.B., Kolupaeva, V.G., Marintchev, A., Wagner, G. & Pestova, T.V. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 17, 2786–2797 (2003).
Johnson, C.H., Kruft, V. & Subramanian, A.R. Identification of a plastid-specific ribosomal protein in the 30S subunit of chloroplast ribosomes and isolation of the cDNA clone encoding its cytoplasmic precursor. J. Biol. Chem. 265, 12790–12795 (1990).
Bubunenko, M.G. & Subramanian, A.R. Recognition of novel and divergent higher plant chloroplast ribosomal proteins by Escherichia coli ribosome during in vivo assembly. J. Biol. Chem. 269, 18223–18231 (1994).
Tan, X., Varughese, M. & Widger, W.R. A light-repressed transcript found in Synechococcus PCC 7002 is similar to a chloroplast-specific small subunit ribosomal protein and to a transcription modulator protein associated with sigma 54. J. Biol. Chem. 269, 20905–20912 (1994).
Otwinowski, Z. & Minor, W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326 (1997).
Brünger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard . Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Kleywegt, G.J. & Read, R.J. Not your average density. Structure 5, 1557–1569 (1997).
Yang, H. et al. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 31, 3450–3460 (2003).
Ramesh, V., Gite, S., Li, Y. & RajBhandary, U.L. Suppressor mutations in Escherichia coli methionyl-tRNA formyltransferase: role of a 16-amino acid insertion module in initiator tRNA recognition. Proc. Natl. Acad. Sci. USA 94, 13524–13529 (1997).
Jelenc, P.C. & Kurland, C.G. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. USA 76, 3174–3178 (1979).
Acknowledgements
We thank J. Holton, G. Meigs and C. Ralston for help with data measurement at the Advanced Light Source. We thank S. Yang for the PY expression clone, D. Wilson for IF expression clones and K. Nierhaus for the tRNAPhe expression plasmid. We also thank J. Doudna, V. Ramakrishnan, M. O'Connor, M. Marletta and A. Dahlberg and his laboratory for helpful comments on the paper. A.V.-S. acknowledges F. Vila for balance and coherence. This work was funded by the US National Institutes of Health and by the Department of Energy.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Vila-Sanjurjo, A., Schuwirth, BS., Hau, C. et al. Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol 11, 1054–1059 (2004). https://doi.org/10.1038/nsmb850
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb850
This article is cited by
-
Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach
Nature Communications (2020)
-
Solution structure of the N-terminal domain of the Staphylococcus aureus hibernation promoting factor
Journal of Biomolecular NMR (2019)
-
Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management
Current Genetics (2018)
-
NMR assignments of the N-terminal domain of Staphylococcus aureus hibernation promoting factor (SaHPF)
Biomolecular NMR Assignments (2018)
-
Large-scale analysis of post-translational modifications in E. coli under glucose-limiting conditions
BMC Genomics (2017)