Abstract
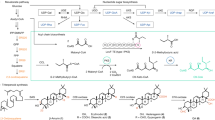
Sulfolipid-1 (SL-1) is an abundant sulfated glycolipid and potential virulence factor found in Mycobacterium tuberculosis. SL-1 consists of a trehalose-2-sulfate (T2S) disaccharide elaborated with four lipids. We identified and characterized a conserved mycobacterial sulfotransferase, Stf0, which generates the T2S moiety of SL-1. Biochemical studies demonstrated that the enzyme requires unmodified trehalose as substrate and is sensitive to small structural perturbations of the disaccharide. Disruption of stf0 in Mycobacterium smegmatis and M. tuberculosis resulted in the loss of T2S and SL-1 formation, respectively. The structure of Stf0 at a resolution of 2.6 Å reveals the molecular basis of trehalose recognition and a unique dimer configuration that encloses the substrate into a bipartite active site. These data provide strong evidence that Stf0 carries out the first committed step in the biosynthesis of SL-1 and establish a system for probing the role of SL-1 in M. tuberculosis infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Mougous, J.D., Green, R.E., Williams, S.J., Brenner, S.E. & Bertozzi, C.R. Sulfotransferases and sulfatases in mycobacteria. Chem. Biol. 9, 767–776 (2002).
Hemmerich, S. & Rosen, S.D. Carbohydrate sulfotransferases in lymphocyte homing. Glycobiology 10, 849–856 (2000).
Honke, K. & Taniguchi, N. Sulfotransferases and sulfated oligosaccharides. Med. Res. Rev. 22, 637–654 (2002).
Moore, K.L. The biology and enzymology of protein tyrosine O-sulfation. J. Biol. Chem. 278, 24243–24246 (2003).
Coughtrie, M.W. Sulfation through the looking glass—recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2, 297–308 (2002).
Hemmerich, S., Butcher, E.C. & Rosen, S.D. Sulfation-dependent recognition of high endothelial venules (HEV)-ligands by L-selectin and MECA-79, and adhesion-blocking monoclonal antibody. J. Exp. Med. 180, 2219–2226 (1994).
Hemmerich, S. et al. Sulfation of L-selectin ligands by an HEV-restricted sulfotransferase regulates lymphocyte homing to lymph nodes. Immunity 15, 237–247 (2001).
Farzan, M. et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676 (1999).
Choe, H. et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114, 161–170 (2003).
Fiete, D., Srivastava, V., Hindsgaul, O. & Baenziger, J.U. A hepatic reticuloendothelial cell receptor specific for SO4-4GalNAcβ 1,4GlcNAcβ 1,2Manα that mediates rapid clearance of lutropin. Cell 67, 1103–1110 (1991).
Hanin, M. et al. Sulphation of Rhizobium sp. NGR234 Nod factors is dependent on noeE, a new host-specificity gene. Mol. Microbiol. 24, 1119–1129 (1997).
Roche, P. et al. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell 67, 1131–1143 (1991).
Shen, Y., Sharma, P., da Silva, F.G. & Ronald, P. The Xanthomonas oryzae pv. lozengeoryzae raxP and raxQ genes encode an ATP sulphurylase and adenosine-5′-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 44, 37–48 (2002).
Mougous, J.D. et al. Discovery of sulfated metabolites in mycobacteria with a genetic and mass spectrometric approach. Proc. Natl. Acad. Sci. USA 99, 17037–17042 (2002).
McCarthy, C. Synthesis and release of sulfolipid by Mycobacterium avium during growth and cell division. Infect. Immun. 14, 1241–1252 (1976).
Khoo, K.H. et al. Altered expression profile of the surface glycopeptidolipids in drug- resistant clinical isolates of Mycobacterium avium complex. J. Biol. Chem. 274, 9778–9785 (1999).
Lopez Marin, L.M. et al. Structure of a novel sulfate-containing mycobacterial glycolipid. Biochemistry 31, 11106–11111 (1992).
Goren, M.B., Brokl, O. & Schaefer, W.B. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: correlation of virulence with elaboration of sulfatides and strongly acidic lipids. Infect. Immun. 9, 142–149 (1974).
Goren, M.B. Sulfolipid I of Mycobacterium tuberculosis, strain H37Rv. I. Purification and properties. Biochim. Biophys. Acta 210, 116–126 (1970).
Daffe, M. & Draper, P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39, 131–203 (1998).
Zhang, L., English, D. & Andersen, B.R. Activation of human neutrophils by Mycobacterium tuberculosis-derived sulfolipid-1. J. Immunol. 146, 2730–2736 (1991).
Zhang, L., Gay, J.C., English, D. & Andersen, B.R. Neutrophil priming mechanisms of sulfolipid-I and N-formyl-methionyl-leucyl-phenylalanine. J. Biomed. Sci. 1, 253–262 (1994).
Zhang, L., Goren, M.B., Holzer, T.J. & Andersen, B.R. Effect of Mycobacterium tuberculosis-derived sulfolipid I on human phagocytic cells. Infect. Immun. 56, 2876–2883 (1988).
Julian, E. et al. Serodiagnosis of tuberculosis: comparison of immunoglobulin A (IgA) response to sulfolipid I with IgG and IgM responses to 2,3-diacyltrehalose, 2,3,6-triacyltrehalose, and cord factor antigens. J. Clin. Microbiol. 40, 3782–3788 (2002).
Julian, E., Matas, L., Alcaide, J. & Luquin, M. Comparison of antibody responses to a potential combination of specific glycolipids and proteins for test sensitivity improvement in tuberculosis serodiagnosis. Clin. Diagn. Lab. Immunol. 11, 70–76 (2004).
Converse, S.E. et al. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. USA 100, 6121–6126 (2003).
Gilleron, M. et al. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J. Exp. Med. 199, 649–659 (2004).
Sirakova, T.D., Thirumala, A.K., Dubey, V.S., Sprecher, H. & Kolattukudy, P.E. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276, 16833–16839 (2001).
Domenech, P. et al. The role of MmpL8 in sulfatide biogensis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279, 21257–21265 (2004).
Rousseau, C. et al. Sulfolipid deficiency does not affect the virulence of Mycobacterium tuberculosis H37Rv in mice and guinea pigs. Infect. Immun. 71, 4684–4690 (2003).
Cronan, G.E. & Keating, D.H. Sinorhizobium meliloti sulfotransferase that modifies lipopolysaccharide. J. Bacteriol. 186, 4168–4176 (2004).
Cole, S.T. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998).
Kakuta, Y., Pedersen, L.G., Pedersen, L.C. & Negishi, M. Conserved structural motifs in the sulfotransferase family. Trends Biochem. Sci. 23, 129–130 (1998).
Elbein, A.D. & Mitchell, M. Levels of glycogen and trehalose in Mycobacterium smegmatis and the purification and properties of the glycogen synthetase. J. Bacteriol. 113, 863–873 (1973).
Petzold, C.J., Leavell, M.D. & Leary, J.A. Screening and identification of acidic carbohydrates in bovine colostrum by using ion/molecule reactions and Fourier transform ion cyclotron resonance mass spectrometry: specificity toward phosphorylated complexes. Anal. Chem. 76, 203–210 (2004).
Negishi, M. et al. Structure and function of sulfotransferases. Arch. Biochem. Biophys. 390, 149–157 (2001).
Edavettal, S.C. et al. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J. Biol. Chem. 279, 25789–25797 (2004).
Kakuta, Y., Sueyoshi, T., Negishi, M. & Pedersen, L.C. Crystal structure of the sulfotransferase domain of human heparan sulfate N-deacetylase/ N-sulfotransferase 1. J. Biol. Chem. 274, 10673–10676 (1999).
Kakuta, Y., Pedersen, L.G., Carter, C.W., Negishi, M. & Pedersen, L.C. Crystal structure of estrogen sulphotransferase. Nat. Struct. Biol. 4, 904–908 (1997).
Petrotchenko, E.V., Pedersen, L.C., Borchers, C.H., Tomer, K.B. & Negishi, M. The dimerization motif of cytosolic sulfotransferases. FEBS Lett. 490, 39–43 (2001).
Goodsell, D.S. & Olson, A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29, 105–153 (2000).
Chapman, E., Bryan, M.C. & Wong, C.H. Mechanistic studies of β-arylsulfotransferase IV. Proc. Natl. Acad. Sci. USA 100, 910–5 (2003).
Kakuta, Y., Petrotchenko, E.V., Pedersen, L.C. & Negishi, M. The sulfuryl transfer mechanism. Crystal structure of a vanadate complex of estrogen sulfotransferase and mutational analysis. J. Biol. Chem. 273, 27325–27330 (1998).
Chapman, E., Bryan, M.C. & Wong, C.H. Mechanistic studies of β-arylsulfotransferase IV. Proc. Natl. Acad. Sci. USA 100, 910–915 (2003).
Pedersen, L.C., Petrotchenko, E., Shevtsov, S. & Negishi, M. Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction. J. Biol. Chem. 277, 17928–17932 (2002).
Rini, J.M. Lectin structure. Annu. Rev. Biophys. Biomol. Struct. 24, 551–577 (1995).
Elbein, A.D., Pan, Y.T., Pastuszak, I. & Carroll, D. New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R–27R (2003).
Wolf, A., Kramer, R. & Morbach, S. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol. Microbiol. 49, 1119–1134 (2003).
Gibson, R.P., Turkenburg, J.P., Charnock, S.J., Lloyd, R. & Davies, G.J. Insights into trehalose synthesis provided by the structure of the retaining glucosyltransferase OtsA. Chem. Biol. 9, 1337–1346 (2002).
Ronning, D.R. et al. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 7, 141–146 (2000).
Armstrong, J.I. et al. A library approach to the generation of bisubstrate analogue sulfotransferase inhibitors. Org. Lett. 3, 2657–2660 (2001).
Verdugo, D.E. et al. Discovery of estrogen sulfotransferase inhibitors from a purine library screen. J. Med. Chem. 44, 2683–2686 (2001).
Snapper, S.B., Melton, R.E., Mustafa, S., Kieser, T. & Jacobs, W.R. Jr. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919 (1990).
Parish, T. & Stoker, N.G. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975 (2000).
Kaps, I. et al. Energy transfer between fluorescent proteins using a co-expression system in Mycobacterium smegmatis. Gene 278, 115–124 (2001).
Harth, G. & Horwitz, M.A. Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J. Biol. Chem. 272, 22728–22735 (1997).
Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276, 307–326 (1997).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Holton, J. & Alber, T. Automated protein crystal structure determination using ELVES. Proc. Natl. Acad. Sci. USA 101, 1537–1542 (2004).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57, 122–133 (2001).
Lamzin, V.S. & Wilson, K.S. Automated refinement of protein models. Acta Crystallogr. D 49, 129–147 (1993).
Ehrhardt, D.W. et al. In vitro sulfotransferase activity of NodH, a nodulation protein of Rhizobium meliloti required for host-specific nodulation. J. Bacteriol. 177, 6237–6245 (1995).
Pratt, M.R., Leigh, C.D. & Bertozzi, C.R. Formation of 1,1-α,α-glycosidic bonds by intramolecular aglycone delivery. A convergent synthesis of trehalose. Org. Lett. 5, 3185–3188 (2003).
Langston, S., Bernet, B. & Vasella, A. Temporary protection and activation in the regioselective synthesis of saccharide sulfates. Helv. Chim. Acta 77, 2341–2353 (1994).
Goren, M.B. Sulfolipid I of Mycobacterium tuberculosis, strain H37Rv. II. Structural studies. Biochim. Biophys. Acta 210, 127–138 (1970).
Minnikin, D.E., Kremer, L., Dover, L.G. & Besra, G.S. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 9, 545–553 (2002).
Acknowledgements
We thank the TB Structural Genomics Consortium (http://www.doe-mbi.ucla.edu/TB) for partial funding. We are grateful to J. Holton and C. Ralston at the Advanced Light Source for their assistance with data collection and processing, D. Chen and M. Schelle for fruitful scientific discussions, members of the Berger and Bertozzi labs for careful review of the manuscript, D. Keating for sharing valuable sequence data and H. Schachman for use of equipment. J.D.M. was supported by a fellowship from the Ford Foundation. This work was supported by a grant from the US National Institutes of Health to C.R.B. (AI51622) and J.M.B. (P50-GM62410).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Multiple sequence alignment of Stf0 with various sulfotransferases. (PDF 426 kb)
Supplementary Figure 2
Analytical equilibrium centrifugation of Stf0 (PDF 116 kb)
Supplementary Table 1
Data collection and refinement statistics. (PDF 42 kb)
Supplementary Table 2
Distances of hydrogen bonds discussed in text. (PDF 30 kb)
Rights and permissions
About this article
Cite this article
Mougous, J., Petzold, C., Senaratne, R. et al. Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis. Nat Struct Mol Biol 11, 721–729 (2004). https://doi.org/10.1038/nsmb802
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb802
This article is cited by
-
Engineering sulfonate group donor regeneration systems to boost biosynthesis of sulfated compounds
Nature Communications (2023)
-
Computational identification of significant immunogenic epitopes of the putative outer membrane proteins from Mycobacterium tuberculosis
Journal of Genetic Engineering and Biotechnology (2021)
-
Candidatus Eremiobacterota, a metabolically and phylogenetically diverse terrestrial phylum with acid-tolerant adaptations
The ISME Journal (2021)
-
The vesicle-associated function of NOD2 as a link between Crohn’s disease and mycobacterial infection
Gut Pathogens (2015)
-
Trehalose/2-sulfotrehalose biosynthesis and glycine-betaine uptake are widely spread mechanisms for osmoadaptation in the Halobacteriales
The ISME Journal (2014)