Abstract
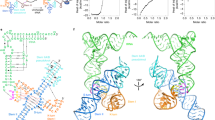
The tandem zinc finger (TZF) domain of the protein TIS11d binds to the class II AU-rich element (ARE) in the 3′ untranslated region (3′ UTR) of target mRNAs and promotes their deadenylation and degradation. The NMR structure of the TIS11d TZF domain bound to the RNA sequence 5′-UUAUUUAUU-3′ comprises a pair of novel CCCH fingers of type CX8CX5CX3H separated by an 18-residue linker. The two TIS11d zinc fingers bind in a symmetrical fashion to adjacent 5′-UAUU-3′ subsites on the single-stranded RNA via a combination of electrostatic and hydrogen-bonding interactions, with intercalative stacking between conserved aromatic side chains and the RNA bases. Sequence specificity in RNA recognition is achieved by a network of intermolecular hydrogen bonds, mostly between TIS11d main-chain functional groups and the Watson-Crick edges of the bases. The TIS11d structure provides insights into the RNA-binding functions of this large family of CCCH zinc finger proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Accessions
GenBank/EMBL/DDBJ
Protein Data Bank
References
Blackshear, P.J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30, 945–952 (2002).
Ming, X.F., Stoecklin, G., Lu, M., Looser, R. & Moroni, C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 21, 5778–5789 (2001).
Varnum, B.C., Ma, Q.F., Chi, T.H., Fletcher, B. & Herschman, H.R. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol. Cell. Biol. 11, 1754–1758 (1991).
Lai, W.S., Stumpo, D.J. & Blackshear, P.J. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J. Biol. Chem. 265, 16556–16563 (1990).
DuBois, R.N., McLane, M.W., Ryder, K., Lau, L.F. & Nathans, D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J. Biol. Chem. 265, 19185–19191 (1990).
Lai, W.S., Carballo, E., Thorn, J.M., Kennington, E.A. & Blackshear, P.J. Interactions of CCCH zinc finger proteins with mRNA-binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275, 17827–17837 (2000).
Carballo, E., Lai, W.S. & Blackshear, P.J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281, 1001–1005 (1998).
Taylor, G.A. et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454 (1996).
Chen, C.Y. et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107, 451–464 (2001).
Lai, W.S., Kennington, E.A. & Blackshear, P.J. Interactions of CCCH zinc finger proteins with mRNA. Non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 277, 9606–9613 (2002).
Worthington, M.T., Amann, B.T., Nathans, D. & Berg, J.M. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc. Natl. Acad. Sci. USA 93, 13754–13759 (1996).
Lai, W.S. et al. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 19, 4311–4323 (1999).
Worthington, M.T. et al. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J. Biol. Chem. 277, 48558–48564 (2002).
Blackshear, P.J. et al. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J. Biol. Chem. 278, 19947 (2003).
De Guzman, R.N. et al. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Psi-RNA recognition element. Science 279, 384–388 (1998).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Amann, B.T., Worthington, M.T. & Berg, J.M. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry 42, 217–221 (2003).
Chen, C.Y. & Shyu, A.B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci 20, 465–470 (1995).
Wang, X. & Tanaka Hall, T.M. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 8, 141–145 (2001).
Handa, N. et al. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398, 579–585 (1999).
Deo, R.C., Bonanno, J.B., Sonenberg, N. & Burley, S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98, 835–845 (1999).
Guedes, S. & Priess, J.R. The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development 124, 731–739 (1997).
Tabara, H., Hill, R.J., Mello, C.C., Priess, J.R. & Kohara, Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1–11 (1999).
Reese, K.J., Dunn, M.A., Waddle, J.A. & Seydoux, G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell 6, 445–455 (2000).
Bai, C.Y. & Tolias, P.P. Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol. Cell. Biol. 16, 6661–6667 (1996).
Hendriks, E.F., Robinson, D.R., Hinkins, M. & Matthews, K.R. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J. 20, 6700–6711 (2001).
Wishart, D.S. et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140 (1995).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Johnson, B.A. & Blevins, R.A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 604–613 (1994).
Wittekind, M. & Mueller, L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. 101, 201–205 (1993).
Grzesiek, S. & Bax, A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114, 6291–6293 (1992).
Grzesiek, S. & Bax, A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J. Magn. Reson. 96, 432–440 (1992).
Grzesiek, S., Anglister, J. & Bax, A. Correlation of backbone amide and aliphatic side chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B 101, 114–119 (1993).
Bax, A., Clore, G.M. & Gronenborn, A.M. 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 88, 425–431 (1990).
Yamazaki, T., Forman-Kay, J.D. & Kay, L.E. Two-dimensional NMR experiments for correlating 13Cβ and 1Hδ/ε chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 115, 11054–11055 (1993).
Vuister, G.W., Delaglio, F. & Bax, A. The use of 1JCαHα coupling constants as a probe for protein backbone conformation. J. Biomol. NMR 3, 67–80 (1993).
Archer, S.J., Ikura, M., Torchia, D.A. & Bax, A. An alternative 3D NMR technique for correlating backbone 15N with side chain Hβ resonances in larger proteins. J. Magn. Reson. 95, 636–641 (1991).
Zwahlen, C. et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage λ N-peptide/boxB RNA complex. J. Am. Chem. Soc. 119, 6711–6721 (1997).
Grzesiek, S., Kuboniwa, H., Hinck, A.P. & Bax, A. Multiple-quantum line narrowing for measurement of Hα-Hβ J coupling in isotopically enriched proteins. J. Am. Chem. Soc. 117, 5312–5315 (1995).
Grzesiek, S., Vuister, G.W. & Bax, A. A simple and sensitive experiment for measurement of JCC couplings between backbone carbonyl and methyl carbons in isotopically enriched proteins. J. Biomol. NMR 3, 487–493 (1993).
Vuister, G.W., Wang, A.C. & Bax, A. Measurement of three-bond nitrogen-carbon J couplings in proteins uniformly enriched in 15N and 13C. J. Am. Chem. Soc. 115, 5334–5335 (1993).
Hansen, M.R., Mueller, L. & Pardi, A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 5, 1065–1074 (1998).
Wang, Y.X. et al. Simultaneous measurement of 1H-15N, 1H-13C', and 15N-13C' dipolar couplings in a perdeuterated 30 kDa protein dissolved in a dilute liquid crystalline phase. J. Am. Chem. Soc. 120, 7385–7386 (1998).
Yang, D., Tolman, J.R., Goto, N.K. & Kay, L.E. An HNCO-based pulse scheme for the measurement of 13Cα-1Hα one-bond dipolar couplings in 15N, 13C labeled proteins. J. Biomol. NMR 12, 325–332 (1998).
Güntert, P., Mumenthaler, C. & Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997).
Duggan, B.M., Legge, G.B., Dyson, H.J. & Wright, P.E. SANE (structure assisted NOE evaluation): an automated model-based approach for NOE assignment. J. Biomol. NMR 19, 321–329 (2001).
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999).
Tsui, V. & Case, D.A. Molecular simulations of nucleic acids using a generalized Born solvation model. J. Am. Chem. Soc. 122, 2489–2498 (2000).
Dosset, P., Hus, J.C., Marion, D. & Blackledge, M. A novel interactive tool for rigid-body modeling of multi-domain macromolecules using residual dipolar couplings. J. Biomol. NMR 20, 223–231 (2001).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 14, 51–55 (1996).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Laskowski, R.A., Rullmann, J.A.C., MacArthur, M.W., Kaptein, R. & Thornton, J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996).
Acknowledgements
We thank B. Lee, M. Hennig, J. Williamson and D. Case for invaluable discussions, J. Chung and G. Kroon for assistance with NMR experiments, and M. Allen and L. Tennant for technical assistance. This work was supported by a grant from the US National Institutes of Health and by the Skaggs Institute for Chemical Biology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Hudson, B., Martinez-Yamout, M., Dyson, H. et al. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11, 257–264 (2004). https://doi.org/10.1038/nsmb738
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb738
This article is cited by
-
Downregulation of Linc00173 increases BCL2 mRNA stability via the miR-1275/PROCA1/ZFP36L2 axis and induces acquired cisplatin resistance of lung adenocarcinoma
Journal of Experimental & Clinical Cancer Research (2023)
-
Towards resolution of the intron retention paradox in breast cancer
Breast Cancer Research (2022)
-
CCCH Zinc finger genes in Barley: genome-wide identification, evolution, expression and haplotype analysis
BMC Plant Biology (2022)
-
Backbone and sidechain 1H, 15N and 13C resonance assignments of the free and RNA-bound tandem zinc finger domain of the tristetraprolin family member from Selaginella moellendorffii
Biomolecular NMR Assignments (2022)
-
Pb(II) coordination to the nonclassical zinc finger tristetraprolin: retained function with an altered fold
JBIC Journal of Biological Inorganic Chemistry (2022)