Abstract
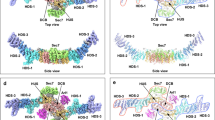
Recruitment of the GRIP domain golgins to the trans-Golgi network is mediated by Arl1, a member of the ARF/Arl small GTPase family, through interaction between their GRIP domains and Arl1-GTP. The crystal structure of Arl1-GTP in complex with the GRIP domain of golgin-245 shows that Arl1-GTP interacts with the GRIP domain predominantly in a hydrophobic manner, with the switch II region conferring the main recognition surface. The involvement of the switch and interswitch regions in the interaction between Arl1-GTP and GRIP accounts for the specificity of GRIP domain for Arl1-GTP. Mutations that abolished the Arl1-mediated Golgi localization of GRIP domain golgins have been mapped on the interface between Arl1-GTP and GRIP. Notably, the GRIP domain forms a homodimer in which each subunit interacts separately with one Arl1-GTP. Mutations disrupting the GRIP domain dimerization also abrogated its Golgi targeting, suggesting that the dimeric form of GRIP domain is a functional unit.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Carter, L.L., Redelmeier, T.E., Woollenweber, L.A. & Schmid, S.L. Multiple GTP-binding proteins participate in clathrin-coated vesicle-mediated endocytosis. J. Cell Biol. 120, 37–45 (1993).
Schwaninger, R., Plutner, H., Bokoch, G.M. & Balch, W.E. Multiple GTP-binding proteins regulate vesicular transport from the ER to Golgi membranes. J. Cell Biol. 119, 1077–1096 (1992).
Boman, A.L. & Kahn, R.A. Arf proteins: the membrane traffic police? Trends Biochem. Sci. 20, 147–150 (1995).
Lowe, S.L., Wong, S.H. & Hong, W. The mammalian ARF-like protein 1 (Arl1) is associated with the Golgi complex. J. Cell Sci. 109, 209–220 (1996).
Schurmann, A. et al. Cloning of two novel ADP-ribosylation factor-like proteins and Characterization of their differential expression in 3T3-L1 cells. J. Biol. Chem. 269, 15683–15688 (1994).
Lu, L., Horstmann, H., Ng, C. & Hong, W.J. Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J. Cell Sci. 114, 4543–4555 (2001).
Van Valkenburgh, H., Shern, J.F., Sharer, J.D., Zhu, X. & Kahn, R.A. ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J. Biol. Chem. 276, 22826–22837 (2001).
Bhamidipati, A., Lewis, S.A. & Cowan, N.J. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149, 1087–1096 (2000).
Fleming, J.A., Vega, L.R. & Solomon, F. Function of tubulin binding proteins in vivo. Genetics 156, 69–80 (2000).
Cuvillier, A. et al. LdARL-3A, a Leishmania promastigote-specific ADP-ribosylation factor-like protein, is essential for flagellum integrity. J. Cell Sci. 113, 2065–2074 (2000).
Antoshechkin, I. & Han, M. The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev. Cell 2, 579–591 (2002).
Tzafrir, I. et al. Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 128, 38–51 (2002).
Grayson, C. et al. Localization in the human retina of the X-linked retinitis pigmentosa protein RP2, its homologue cofactor C and the RP2 interacting protein Arl3. Hum. Mol. Genet. 11, 3065–3074 (2002).
Sharer, J.D. & Kahn, R.A. The ARF-like 2 (ARL2)-binding protein, BART. Purification, cloning, and initial characterization. J. Biol. Chem. 274, 27553–27561 (1999).
Ingley, E. et al. A novel ADP-ribosylation like factor (ARL-6), interacts with the protein-conducting channel SEC61β subunit. FEBS Lett. 459, 69–74 (1999).
Jacobs, S. et al. ADP-ribosylation factor (ARF)-like 4, 6, and 7 represent a subgroup of the ARF family characterization by rapid nucleotide exchange and a nuclear localization signal. FEBS Lett. 456, 384–388 (1999).
Lin, C.Y. et al. ARL4, an ARF-like protein that is developmentally regulated and localized to nuclei and nucleoli. J. Biol. Chem. 275, 37815–37823 (2000).
Schurmann, A. et al. Reduced sperm count and normal fertility in male mice with targeted disruption of the ADP-ribosylation factor-like 4 (Arl4) gene. Mol. Cell Biol. 22, 2761–2768 (2002).
Lin, C.Y., Li, C.C., Huang, P.H. & Lee, F.J. A developmentally regulated ARF-like 5 protein (ARL5), localized to nuclei and nucleoli, interacts with heterochromatin protein 1. J. Cell Sci. 115, 4433–4445 (2002).
Buttitta, L., Tanaka, T.S., Chen, A.E., Ko, M.S. & Fan, C.M. Microarray analysis of somitogenesis reveals novel targets of different WNT signaling pathways in the somitic mesoderm. Dev. Biol. 258, 91–104 (2003).
Munro, S. & Nichols, B.J. The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 9, 377–380 (1999).
Kjer-Nielsen, L., Teasdale, R.D., van Vliet, C. & Gleeson, P.A. A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr. Biol. 9, 385–388 (1999).
Barr, F.A. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol. 9, 381–384 (1999).
Luke, M.R., Kjer-Nielsen, L., Brown, D.L., Stow, J.L. & Gleeson, P.A. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to sub-compartments of the trans-Golgi network. J. Biol. Chem. 278, 4216–4226 (2003).
Gillingham, A.K. & Munro, S. Long coiled-coil proteins and membrane traffic. Biochim. Biophys. Acta 1641, 71–85 (2003).
Brown, D.L. et al. The GRIP domain is a specific targeting sequence for a population of trans-Golgi network derived tubulo-vesicular carriers 1. Traffic 2, 336–344 (2001).
Kjer-Nielsen, L., van Vliet, C., Erlich, R., Toh, B.H. & Gleeson, P.A. The Golgi-targeting sequence of the peripheral membrane protein p230. J. Cell Sci. 112, 1645–1654 (1999).
Lu, L. & Hong, W. Interaction of Arl1-GTP with GRIP domains recruits auto- antigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol. Biol. Cell 14, 3767–3781 (2003).
Panic, B., Whyte, J.R. & Munro, S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 13, 405–410 (2003).
Setty, S.R., Shin, M.E., Yoshino, A., Marks, M.S. & Burd, C.G. Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr. Biol. 13, 401–404 (2003).
Jackson, C.L. Membrane traffic: Arl GTPases get a GRIP on the Golgi. Curr. Biol. 13, R174–176 (2003).
Paduch, M., Jelen, F. & Otlewski, J. Structure of small G proteins and their regulators. Acta Biochim Pol. 48, 829–850 (2001).
Amor, J.C. et al. Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases. J. Biol. Chem. 276, 42477–42484 (2001).
Hillig, R.C. et al. Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein. Struct. Fold Des. 8, 1239–1245 (2000).
Hanzal-Bayer, M., Renault, L., Roversi, P., Wittinghofer, A. & Hillig, R.C. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J. 21, 2095–2106 (2002).
Amor, J.C., Harrison, D.H., Kahn, R.A. & Ringe, D. Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature 372, 704–708 (1994).
Greasley, S.E. et al. The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms. Nat. Struct. Biol. 2, 797–806 (1995).
Goldberg, J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 95, 237–248 (1998).
Pasqualato, S., Menetrey, J., Franco, M. & Cherfils, J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2, 234–238 (2001).
Collins, B.M., Watson, P.J. & Owen, D.J. The structure of the GGA1-GAT domain reveals the molecular basis for ARF binding and membrane association of GGAs. Dev. Cell 4, 321–432 (2003).
Suer, S., Misra, S., Saidi, L.F. & Hurley, J.H. Structure of the GAT domain of human GGA1: a syntaxin amino-terminal domain fold in an endosomal trafficking adaptor. Proc. Natl. Acad. Sci. 100, 4451–4456 (2003).
Shiba, T. et al. Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat. Struct. Biol. 10, 386–393 (2003).
Otwinowski, Z. In Data Collection and Processing (eds. Sawyer, N.I.L. & Bailey, S.) 56–62 (SERC Daresbury Laboratory, Warrington, UK, 1993).
Navaza, J. & Saludjian, P. AMoRe: an automated molecular replacement program package. Methods Enzymol. 276, 581–594 (1997).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000).
Jones, T.A. & Kjeldgaard, M. Electron-density map interpretation. Methods Enzymol. 277, 173–208 (1997).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Murshudov, G.N, Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Gouet, P. & Courcelle, E. ENDscript: a workflow to display sequence and structure information. Bioinformatics 18, 767–768 (2002).
Acknowledgements
We thank P. Tucker at BW7A (European Molecular Biology Laboratory, Hamburg, Germany) for assistance and access to synchrotron radiation facilities, and M.J. Fritzler and E.K.L. Chan for providing the full-length cDNA of human golgin-97. This work is financially supported by the Agency for Science, Technology and Research (A*STAR) in Singapore (to W.H. and H.S.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Wu, M., Lu, L., Hong, W. et al. Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat Struct Mol Biol 11, 86–94 (2004). https://doi.org/10.1038/nsmb714
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb714
This article is cited by
-
The inner workings of intracellular heterotypic and homotypic membrane fusion mechanisms
Journal of Biosciences (2019)
-
Mammalian Mon2/Ysl2 regulates endosome-to-Golgi trafficking but possesses no guanine nucleotide exchange activity toward Arl1 GTPase
Scientific Reports (2013)
-
The structural basis of Arf effector specificity: the crystal structure of ARF6 in a complex with JIP4
The EMBO Journal (2009)


