Abstract
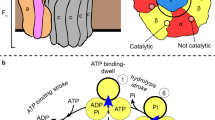
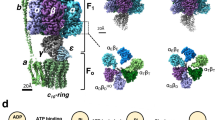
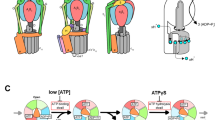
F1-ATPase, the catalytic part of FoF1-ATP synthase, rotates the central γ subunit within the α3β3 cylinder in 120° steps, each step consuming a single ATP molecule. However, how the catalytic activity of each β subunit is coordinated with the other two β subunits to drive rotation remains unknown. Here we show that hybrid F1 containing one or two mutant β subunits with altered catalytic kinetics rotates in an asymmetric stepwise fashion. Analysis of the rotations reveals that for any given β subunit, the subunit binds ATP at 0°, cleaves ATP at ∼200° and carries out a third catalytic event at ∼320°. This demonstrates the concerted nature of the F1 complex activity, where all three β subunits participate to drive each 120° rotation of the γ subunit with a 120° phase difference, a process we describe as a 'sequential three-site mechanism'.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
17 September 2007
figure 4 (b) replaced
Notes
*NOTE: In the version of this article initially published, the proposed model for coupling the rotation and catalysis of the F1 β subunits to the concerted activity of their active sites shown in Figure 4 was incorrectly drawn. The error has been corrected in the HTML and PDF versions of the article
References
Boyer, P.D. The ATP synthase–a splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 (1997).
Kinosita, K., Jr., Adachi, K. & Itoh, H. Rotation of F1-ATPase: how an ATP-driven molecular machine may work. Annu. Rev. Biophys. Biomol. Struct. 33, 245–268 (2004).
Kayalar, C., Rosing, J. & Boyer, P.D. An alternating site sequence for oxidative phosphorylation suggested by measurement of substrate binding patterns and exchange reaction inhibitions. J. Biol. Chem. 252, 2486–2491 (1977).
Gresser, M.J., Myers, J.A. & Boyer, P.D. Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model. J. Biol. Chem. 257, 12030–12038 (1982).
Grubmeyer, C., Cross, R.L. & Penefsky, H.S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase. Rate constants for elementary steps in catalysis at a single site. J. Biol. Chem. 257, 12092–12100 (1982).
Cross, R.L., Grubmeyer, C. & Penefsky, H.S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase. Rate enhancements resulting from cooperative interactions between multiple catalytic sites. J. Biol. Chem. 257, 12101–12105 (1982).
Boyer, P.D. The binding change mechanism for ATP synthase–some probabilities and possibilities. Biochim. Biophys. Acta 1140, 215–250 (1993).
Abrahams, J.P., Leslie, A.G., Lutter, R. & Walker, J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994).
Bowler, M.W., Montgomery, M.G., Leslie, A.G. & Walker, J.E. How azide inhibits ATP hydrolysis by the F-ATPases. Proc. Natl. Acad. Sci. USA 103, 8646–8649 (2006).
Bowler, M.W., Montgomery, M.G., Leslie, A.G. & Walker, J.E. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 282, 14238–14242 (2007).
Weber, J., Wilke-Mounts, S., Lee, R.S., Grell, E. & Senior, A.E. Specific placement of tryptophan in the catalytic sites of Escherichia coli F1-ATPase provides a direct probe of nucleotide binding: maximal ATP hydrolysis occurs with three sites occupied. J. Biol. Chem. 268, 20126–20133 (1993).
Ren, H., Bandyopadhyay, S. & Allison, W.S. The α3(βMet222Ser/Tyr345Trp)3γ subcomplex of the TF1-ATPase does not hydolyze ATP at a significant rate until the substrate binds to the catalytic site of the lowest affinity. Biochemistry 45, 6222–6230 (2006).
Senior, A.E., Nadanaciva, S. & Weber, J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta 1553, 188–211 (2002).
Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K., Jr. Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 (1997).
Yasuda, R., Noji, H., Kinosita, K., Jr. & Yoshida, M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell 93, 1117–1124 (1998).
Yasuda, R., Noji, H., Yoshida, M., Kinosita, K., Jr. & Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001).
Shimabukuro, K. et al. Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. USA 100, 14731–14736 (2003).
Xing, J., Liao, J.C. & Oster, G. Making ATP. Proc. Natl. Acad. Sci. USA 102, 16539–16546 (2005).
Gao, Y.Q., Yang, W. & Karplus, M. A structure-based model for the synthesis and hydrolysis of ATP by F1-ATPase. Cell 123, 195–205 (2005).
Ariga, T., Masaike, T., Noji, H. & Yoshida, M. Stepping rotation of F1-ATPase with one, two, or three altered catalytic sites that bind ATP only slowly. J. Biol. Chem. 277, 24870–24874 (2002).
Amano, T., Tozawa, K., Yoshida, M. & Murakami, H. Spatial precision of a catalytic carboxylate of F1-ATPase beta subunit probed by introducing different carboxylate-containing side chains. FEBS Lett. 348, 93–98 (1994).
Dittrich, M., Hayashi, S. & Schulten, K. On the mechanism of ATP hydrolysis in F1-ATPase. Biophys. J. 85, 2253–2266 (2003).
Dittrich, M., Hayashi, S. & Schulten, K. ATP hydrolysis in the βTP and βDP catalytic sites of F1-ATPase. Biophys. J. 87, 2954–2967 (2004).
Nishizaka, T. et al. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 11, 142–148 (2004).
Senior, A.E. & Weber, J. Happy motoring with ATP synthase. Nat. Struct. Mol. Biol. 11, 110–112 (2004).
Weber, J., Bowman, C. & Senior, A.E. Specific tryptophan substitution in catalytic sites of Escherichia coli F1-ATPase allows differentiation between bound substrate ATP and product ADP in steady-state catalysis. J. Biol. Chem. 271, 18711–18718 (1996).
Weber, J. & Senior, A.E. Effects of the inhibitors azide, dicyclohexylcarbodiimide, and aurovertin on nucleotide binding to the three F1-ATPase catalytic sites measured using specific tryptophan probes. J. Biol. Chem. 273, 33210–33215 (1998).
Schlichting, I. et al. Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature 345, 309–315 (1990).
Antes, I., Chandler, D., Wang, H. & Oster, G. The unbinding of ATP from F1-ATPase. Biophys. J. 85, 695–706 (2003).
Rosing, J., Kayalar, C. & Boyer, P.D. Evidence for energy-dependent change in phosphate binding for mitochondrial oxidative phosphorylation based on measurements of medium and intermediate phosphate-water exchanges. J. Biol. Chem. 252, 2478–2485 (1977).
Masaike, T., Muneyuki, E., Noji, H., Kinosita, K., Jr. & Yoshida, M. F1-ATPase changes its conformations upon phosphate release. J. Biol. Chem. 277, 21643–21649 (2002).
Monod, J., Wyman, J. & Changeux, J.P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 (1965).
Koshland, D.E., Jr., Nemethy, G. & Filmer, D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385 (1966).
Itoh, H. et al. Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468 (2004).
Rondelez, Y. et al. Highly coupled ATP synthesis by F1-ATPase single molecules. Nature 433, 773–777 (2005).
Kaseda, K., Higuchi, H. & Hirose, K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat. Cell Biol. 5, 1079–1082 (2003).
Asbury, C.L., Fehr, A.N. & Block, S.M. Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302, 2130–2134 (2003).
Yildiz, A. et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 (2003).
Park, H. et al. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol. Cell 21, 331–336 (2006).
Tomishige, M., Klopfenstein, D.R. & Vale, R.D. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science 297, 2263–2267 (2002).
Kitamura, K., Tokunaga, M., Esaki, S., Iwane, A.H. & Yanagida, T. Mechanism of muscle contraction based on stochastic properties of single actomyosin motors observed in vitro. Biophysics 1, 1–19 (2005).
Press, W.H., Teukolsky, S.A., Vetterling, W.T. & Flannery, B.P. Numerical Recipes in C (Cambridge University Press, New York, 1992).
Acknowledgements
We thank P.D. Boyer, K. Kinosita Jr., T. Kodama, H. Itoh, H. Noji, Y. Taniguchi and our other colleagues for valuable discussions, K. Shimabukuro (Tokyo Institute of Technology) for providing the plasmid fragments encoding β(E190D), K. Adachi (Waseda University) and R. Yasuda (Duke University) for programming the image-processing software, and P. Karagiannis for carefully revising the manuscript. This work was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (14380313; to E.M.), and T.A. was supported by a Research Fellowship of the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Contributions
T.A. performed all experiments and data analysis. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Table 1, Supplementary Methods (PDF 171 kb)
Rights and permissions
About this article
Cite this article
Ariga, T., Muneyuki, E. & Yoshida, M. F1-ATPase rotates by an asymmetric, sequential mechanism using all three catalytic subunits. Nat Struct Mol Biol 14, 841–846 (2007). https://doi.org/10.1038/nsmb1296
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1296
This article is cited by
-
Temperature-dependent regulation of electron transport and ATP synthesis in chloroplasts in vitro and in silico
Photosynthesis Research (2020)
-
Structure and dynamics of rotary V1 motor
Cellular and Molecular Life Sciences (2018)
-
ATP hydrolysis assists phosphate release and promotes reaction ordering in F1-ATPase
Nature Communications (2015)
-
Characterization of the temperature-sensitive reaction of F1-ATPase by using single-molecule manipulation
Scientific Reports (2014)
-
Timing of inorganic phosphate release modulates the catalytic activity of ATP-driven rotary motor protein
Nature Communications (2014)