Abstract
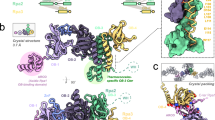
The eukaryotic GINS complex is essential for the establishment of DNA replication forks and replisome progression. We report the crystal structure of the human GINS complex. The heterotetrameric complex adopts a pseudo symmetrical layered structure comprising two heterodimers, creating four subunit-subunit interfaces. The subunit structures of the heterodimers consist of two alternating domains. The C-terminal domains of the Sld5 and Psf1 subunits are connected by linker regions to the core complex, and the C-terminal domain of Sld5 is important for core complex assembly. In contrast, the C-terminal domain of Psf1 does not contribute to the stability of the complex but is crucial for chromatin binding and replication activity. These data suggest that the core complex ensures a stable platform for the C-terminal domain of Psf1 to act as a key interaction interface for other proteins in the replication-initiation process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Blow, J.J. & Dutta, A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6, 476–486 (2005).
Takahashi, T.S., Wigley, D.B. & Walter, J.C. Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem. Sci. 30, 437–444 (2005).
Kanemaki, M., Sanchez-Diaz, A., Gambus, A. & Labib, K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423, 720–724 (2003).
Takayama, Y. et al. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17, 1153–1165 (2003).
Kubota, Y. et al. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 17, 1141–1152 (2003).
Makarova, K.S., Wolf, Y.I., Mekhedov, S.L., Mirkin, B.G. & Koonin, E.V. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 33, 4626–4638 (2005).
Gambus, A. et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 (2006).
Pacek, M., Tutter, A.V., Kubota, Y., Takisawa, H. & Walter, J.C. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21, 581–587 (2006).
Moyer, S.E., Lewis, P.W. & Botchan, M.R. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 103, 10236–10241 (2006).
Marinsek, N. et al. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 7, 539–545 (2006).
De Falco, M. et al. The human GINS complex binds to and specifically stimulates human DNA polymerase alpha-primase. EMBO Rep. 8, 99–103 (2007).
Aparicio, T., Ibarra, A. & Mendez, J. Cdc45-MCM-GINS, a new power player for DNA replication. Cell Div. 1, 18 (2006).
Holm, L. & Sander, C. Alignment of three-dimensional protein structures: network server for database searching. Methods Enzymol. 266, 653–662 (1996).
Grum, V.L., Li, D., MacDonald, R.I. & Mondragon, A. Structures of two repeats of spectrin suggest models of flexibility. Cell 98, 523–535 (1999).
Gomez, E.B., Angeles, V.T. & Forsburg, S.L. A screen for Schizosaccharomyces pombe mutants defective in rereplication identifies new alleles of rad4+, cut9+ and psf2+. Genetics 169, 77–89 (2005).
Jones, S. & Thornton, J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93, 13–20 (1996).
Fukui, T. et al. Distinct roles of DNA polymerases delta and epsilon at the replication fork in Xenopus egg extracts. Genes Cells 9, 179–191 (2004).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Weeks, C.M. & Miller, R. Optimizing Shake-and-Bake for proteins. Acta Crystallogr. D Biol. Crystallogr. 55, 492–500 (1999).
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Chong, J.P., Thommes, P., Rowles, A., Mahbubani, H.M. & Blow, J.J. Characterization of the Xenopus replication licensing system. Methods Enzymol. 283, 549–564 (1997).
Murray, A.W. Cell cycle extracts. Methods Cell Biol. 36, 581–605 (1991).
Mimura, S., Masuda, T., Matsui, T. & Takisawa, H. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 5, 439–452 (2000).
Fiser, A. & Sali, A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 (2003).
Acknowledgements
We thank K. Demura and N. Igarashi at the BL-5A beamline of the Photon Factory for assistance with data collection, M. Usui for mass spectrometry measurements, I. Hayashi, T. Hirano and W. Yang for critical reading of the manuscript and M. Izumi, M. Kanemaki, K. Kimura, S. Tada, A. Takemoto, H. Takisawa, K. Yanagi and Y. Zhiying for helpful comments and discussions. The antibody to Xenopus Pol ε p60 was a gift from S. Waga (Osaka University). This work was supported by Grants-in-Aid for Science Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (K.K.) and in part by Solution Oriented Research for Science and Technology from the Japan Science and Technology Agency (K.K. and F.H.).
Author information
Authors and Affiliations
Contributions
K.K. contributed to structural and molecular biology, biochemistry, manuscript preparation and project direction. Y.K. contributed to molecular biology and manuscript preparation. Y.S. and T.A. performed electron microscopy. F.H. organized the project and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Sequence alignments of GINS subunits. (PDF 445 kb)
Supplementary Fig. 2
Representative electron density map. (PDF 1887 kb)
Supplementary Fig. 3
Superimpositions of A and B domains. (PDF 504 kb)
Supplementary Fig. 4
Coimmunoprecipitation of GINS with other DNA replication proteins. (PDF 308 kb)
Supplementary Data 1
Structural interpretations of other yeast GINS mutants. (PDF 878 kb)
Supplementary Data 2
Electron micrographs and cut-open surface of human GINS complex. (PDF 934 kb)
Rights and permissions
About this article
Cite this article
Kamada, K., Kubota, Y., Arata, T. et al. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat Struct Mol Biol 14, 388–396 (2007). https://doi.org/10.1038/nsmb1231
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1231
This article is cited by
-
Identification of GINS1 as a therapeutic target in the cancer patients infected with COVID-19: a bioinformatics and system biology approach
Hereditas (2022)
-
GINS4 might be a novel prognostic immune-related biomarker of not only esophageal squamous cell carcinoma and other cancers
BMC Medical Genomics (2022)
-
Gene expression profiling and protein–protein interaction analysis reveals the dynamic role of MCM7 in Alzheimer's disorder and breast cancer
3 Biotech (2022)
-
Differential expression of lung adenocarcinoma transcriptome with signature of tobacco exposure
Journal of Applied Genetics (2020)
-
LSH interacts with and stabilizes GINS4 transcript that promotes tumourigenesis in non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2019)