Abstract
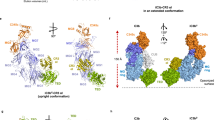
Factor B is the central protease of the complement system of immune defense. Here, we present the crystal structure of human factor B at 2.3-Å resolution, which reveals how the five-domain proenzyme is kept securely inactive. The canonical activation helix of the Von Willebrand factor A (VWA) domain is displaced by a helix from the preceding domain linker. The two helices conformationally link the scissile-activation peptide and the metal ion–dependent adhesion site required for binding of the ligand C3b. The data suggest that C3b binding displaces the three N-terminal control domains and reshuffles the two central helices. Reshuffling of the helices releases the scissile bond for final proteolytic activation and generates a new interface between the VWA domain and the serine protease domain. This allosteric mechanism is crucial for tight regulation of the complement-amplification step in the immune response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Carroll, M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 5, 981–986 (2004).
Walport, M.J. Complement. First of two parts. N. Engl. J. Med. 344, 1058–1066 (2001).
Fishelson, Z., Pangburn, M.K. & Muller-Eberhard, H.J. Characterization of the initial C3 convertase of the alternative pathway of human complement. J. Immunol. 132, 1430–1434 (1984).
Xu, Y., Narayana, S.V. & Volanakis, J.E. Structural biology of the alternative pathway convertase. Immunol. Rev. 180, 123–135 (2001).
Pangburn, M.K. & Muller-Eberhard, H.J. The C3 convertase of the alternative pathway of human complement. Enzymic properties of the bimolecular proteinase. Biochem. J. 235, 723–730 (1986).
Pryzdial, E.L. & Isenman, D.E. Alternative complement pathway activation fragment Ba binds to C3b. Evidence that formation of the factor B-C3b complex involves two discrete points of contact. J. Biol. Chem. 262, 1519–1525 (1987).
Horiuchi, T., Macon, K.J., Engler, J.A. & Volanakis, J.E. Site-directed mutagenesis of the region around Cys-241 of complement component C2. Evidence for a C4b binding site. J. Immunol. 147, 584–589 (1991).
Emsley, J., Knight, C.G., Farndale, R.W., Barnes, M.J. & Liddington, R.C. Structural basis of collagen recognition by integrin alpha2beta1. Cell 101, 47–56 (2000).
Springer, T.A. Complement and the multifaceted functions of VWA and integrin I domains. Structure 14, 1611–1616 (2006).
Ponnuraj, K. et al. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol. Cell 14, 17–28 (2004).
Milder, F.J. et al. Structure of complement component C2a: implications for convertase formation and substrate binding. Structure 14, 1587–1597 (2006).
Smith, C.A., Vogel, C.W. & Muller-Eberhard, H.J. MHC Class III products: an electron microscopic study of the C3 convertases of human complement. J. Exp. Med. 159, 324–329 (1984).
Shimaoka, M. et al. Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell 112, 99–111 (2003).
Bhattacharya, A.A., Lupher, M.L., Jr., Staunton, D.E. & Liddington, R.C. Crystal structure of the A domain from complement factor B reveals an integrin-like open conformation. Structure 12, 371–378 (2004).
Emsley, J., King, S.L., Bergelson, J.M. & Liddington, R.C. Crystal structure of the I domain from integrin alpha2beta1. J. Biol. Chem. 272, 28512–28517 (1997).
Fishelson, Z., Pangburn, M.K. & Muller-Eberhard, H.J. C3 convertase of the alternative complement pathway. Demonstration of an active, stable C3b, Bb (Ni) complex. J. Biol. Chem. 258, 7411–7415 (1983).
Hourcade, D.E., Mitchell, L., Kuttner-Kondo, L.A., Atkinson, J.P. & Medof, M.E. Decay-accelerating factor (DAF), complement receptor 1 (CR1), and factor H dissociate the complement AP C3 convertase (C3bBb) via sites on the type A domain of Bb. J. Biol. Chem. 277, 1107–1112 (2002).
Hourcade, D.E., Mitchell, L.M. & Oglesby, T.J. Mutations of the type A domain of complement factor B that promote high-affinity C3b-binding. J. Immunol. 162, 2906–2911 (1999).
Williams, S.C., Hinshelwood, J., Perkins, S.J. & Sim, R.B. Production and functional activity of a recombinant von Willebrand factor-A domain from human complement factor B. Biochem. J. 342, 625–632 (1999).
Kam, C.M. et al. Human complement proteins D, C2, and B. Active site mapping with peptide thioester substrates. J. Biol. Chem. 262, 3444–3451 (1987).
Khan, A.R. & James, M.N. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 7, 815–836 (1998).
Hourcade, D.E., Wagner, L.M. & Oglesby, T.J. Analysis of the short consensus repeats of human complement factor B by site-directed mutagenesis. J. Biol. Chem. 270, 19716–19722 (1995).
Xu, Y. & Volanakis, J.E. Contribution of the complement control protein modules of C2 in C4b binding assessed by analysis of C2/factor B chimeras. J. Immunol. 158, 5958–5965 (1997).
Thurman, J.M. et al. A novel inhibitor of the alternative complement pathway prevents antiphospholipid antibody-induced pregnancy loss in mice. Mol. Immunol. 42, 87–97 (2005).
Reeves, P.J., Callewaert, N., Contreras, R. & Khorana, H.G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA 99, 13419–13424 (2002).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Storoni, L.C., McCoy, A.J. & Read, R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 (2004).
Jing, H. et al. New structural motifs on the chymotrypsin fold and their potential roles in complement factor B. EMBO J. 19, 164–173 (2000).
Perrakis, A., Morris, R. & Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 (1999).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Terwilliger, T.C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 (2003).
Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 (2001).
Janssen, B.J., Christodoulidou, A., McCarthy, A., Lambris, J.D. & Gros, P. Structure of C3b reveals conformational changes that underlie complement activity. Nature 444, 213–216 (2006).
Wiesmann, C. et al. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature 444, 217–220 (2006).
Acknowledgements
We thank the European Synchrotron Radiation Facility for providing synchrotron radiation facilities and the beamline scientists at ID-14-EH4 for their help with data collection. This work was supported by a 'Pionier' program grant (P.G.) of the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Stereo figure showing electron density (PDF 695 kb)
Supplementary Fig. 2
Activation state of the VWA domain (PDF 299 kb)
Supplementary Fig. 3
The serine protease domain catalytic center (PDF 177 kb)
Supplementary Fig. 4
The CCP triad arrangementF (PDF 400 kb)
Supplementary Fig. 5
The Mg2+-dependent C3b-binding site (PDF 366 kb)
Supplementary Table 1
Analysis of CCP domains 1–3 (PDF 37 kb)
Supplementary Table 2
Contacts between domains CCP1–CCP3 and the VWA and SP domains (PDF 43 kb)
Rights and permissions
About this article
Cite this article
Milder, F., Gomes, L., Schouten, A. et al. Factor B structure provides insights into activation of the central protease of the complement system. Nat Struct Mol Biol 14, 224–228 (2007). https://doi.org/10.1038/nsmb1210
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1210
This article is cited by
-
Novel Functional Assay to Characterize Mutations in Alternative Pathway of Complement
Journal of Clinical Immunology (2023)
-
Insight into mode-of-action and structural determinants of the compstatin family of clinical complement inhibitors
Nature Communications (2022)
-
How novel structures inform understanding of complement function
Seminars in Immunopathology (2018)
-
The properdin pathway: an “alternative activation pathway” or a “critical amplification loop” for C3 and C5 activation?
Seminars in Immunopathology (2018)
-
Molecular mechanisms of complement evasion: learning from staphylococci and meningococci
Nature Reviews Microbiology (2010)