Abstract
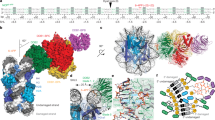
DNA-damage recognition in the nucleotide excision repair (NER) cascade is a complex process, operating on a wide variety of damages. UvrB is the central component in prokaryotic NER, directly involved in DNA-damage recognition and guiding the DNA through repair synthesis. We report the first structure of a UvrB–double-stranded DNA complex, providing insights into the mechanism by which UvrB binds DNA, leading to formation of the preincision complex. One DNA strand, containing a 3′ overhang, threads behind a β-hairpin motif of UvrB, indicating that this motif inserts between the strands of the double helix, thereby locking down either the damaged or undamaged strand. The nucleotide directly behind the β-hairpin is flipped out and inserted into a small, highly conserved pocket in UvrB.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Friedberg, E.C., Walker, G.C. & Siede, W. DNA repair and mutagenesis. (ASM Press, Washington, D.C., 1995).
Sancar, A. DNA excision repair. Annu. Rev. Biochem. 65, 43–81 (1996).
Lloyd, R.S. & Van Houten, B . DNA damage recognition. in DNA Repair Mechanisms: Impact on Human Diseases and Cancer (ed. Vos, J.-M.) 25–66 (R.G. Landes Company, Biomedical Publishers, Austin, Texas, 1995).
Goosen, N. & Moolenaar, G.F. Role of ATP hydrolysis by UvrA and UvrB during nucleotide excision repair. Res. Microbiol. 152, 401–409 (2001).
Van Houten, B. Nucleotide excision repair in Escherichia coli. Microbiol. Rev. 54, 18–51 (1990).
Theis, K. et al. The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch. Mutat. Res. 460, 277–300 (2000).
Verhoeven, E.E., Wyman, C., Moolenaar, G.F. & Goosen, N. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. EMBO J. 21, 4196–4205 (2002).
Orren, D.K. & Sancar, A. Formation and enzymatic properties of the UvrB-DNA complex. J. Biol. Chem. 265, 15796–15803 (1990).
Skorvaga, M., Theis, K., Mandavilli, B.S., Kisker, C. & Van Houten, B. The beta-hairpin motif of UvrB is essential for DNA binding, damage processing, and UvrC-mediated incisions. J. Biol. Chem. 277, 1553–1559 (2002).
Lin, J.-J. & Sancar, A. Active site of (A)BC excinuclease: I. Evidence for 5′ incision by UvrC through a catalytic site involving Asp399, Asp438, and His538 residues. J. Biol. Chem. 267, 17688–17692 (1992).
Sancar, A. & Rupp, W.D. A novel repair enzyme: UvrABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33, 249–260 (1983).
Verhoeven, E.E., van Kesteren, M., Moolenaar, G.F., Visse, R. & Goosen, N. Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J. Biol. Chem. 275, 5120–5123 (2000).
Caron, P.R., Kushner, S.R. & Grossman, L. Involvement of helicase-II (UvrD gene product) and DNA Polymerase-I in excision mediated by the UvrABC protein complex. Proc. Natl. Acad. Sci. USA 82, 4925–4929 (1985).
Husain, I., Houten, B.V., Thomas, D.C., Abdel-Monem, M. & Sancar, A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc. Natl. Acad. Sci. USA 82, 6774–6778 (1985).
Machius, M., Henry, L., Palnitkar, M. & Deisenhofer, J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl. Acad. Sci. USA 96, 11717–11722 (1999).
Nakagawa, N. et al. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem. 126, 986–990 (1999).
Theis, K., Chen, P.J., Skorvaga, M., Houten, B.V. & Kisker, C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 18, 6899–6907 (1999).
Truglio, J.J. et al. Interactions between UvrA and UvrB: the role of UvrB's domain 2 in nucleotide excision repair. EMBO J. 23, 2498–2509 (2004).
Hsu, D.S., Kim, S.T., Sun, Q. & Sancar, A. Structure and function of the UvrB protein. J. Biol. Chem. 270, 8319–8327 (1995).
Gordienko, I. & Rupp, W.D. The limited strand-separating activity of the UvrAB protein complex and its role in the recognition of DNA damage. EMBO J. 16, 889–895 (1997).
Visse, R., King, A., Moolenaar, G.F., Goosen, N. & van de Putte, P. Protein-DNA interactions and alterations in the DNA structure upon UvrB-DNA preincision complex formation during nucleotide excision repair in Escherichia coli. Biochemistry 33, 9881–9888 (1994).
Zou, Y. & Van Houten, B. Strand opening by the UvrA2B complex allows dynamic recognition of DNA damage. EMBO J. 18, 4889–4901 (1999).
Skorvaga, M. et al. Identification of residues within UvrB that are important for efficient DNA binding and damage processing. J. Biol. Chem. 279, 51574–51580 (2004).
DellaVecchia, M.J. et al. Analyzing the handoff of DNA from UvrA to UvrB utilizing DNA-protein photoaffinity labeling. J. Biol. Chem. 279, 45245–45256 (2004).
Moolenaar, G.F., Hoglund, L. & Goosen, N. Clue to damage recognition by UvrB: residues in the beta-hairpin structure prevent binding to non-damaged DNA. EMBO J. 20, 6140–6149 (2001).
Moolenaar, G.F., Schut, M. & Goosen, N. Binding of the UvrB dimer to non-damaged and damaged DNA: residues Y92 and Y93 influence the stability of both subunits. DNA Repair (Amst.) 4, 699–713 (2005).
Zou, Y. et al. DNA damage recognition of mutated forms of UvrB proteins in nucleotide excision repair. Biochemistry 43, 4196–4205 (2004).
Sancar, A., Franklin, K.A. & Sancar, G.B. Escherichia coli DNA photolyase stimulates uvrABC excision nuclease in vitro. Proc. Natl. Acad. Sci. USA 81, 7397–7401 (1984).
Mees, A. et al. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science 306, 1789–1793 (2004).
Malta, E., Moolenaar, G.F. & Goosen, N. Base flipping in nucleotide excision repair. J. Biol. Chem. 281, 2184–2194 (2006).
Zou, Y., Walker, R., Bassett, H., Geacintov, N.E. & Houten, B.V. Formation of DNA repair intermediates and incision by the ATP-dependent UvrB-UvrC endonuclease. J. Biol. Chem. 272, 4820–4827 (1997).
Moolenaar, G.F. et al. The effect of the DNA flanking the lesion on formation of the UvrB-DNA preincision complex. J. Biol. Chem. 275, 8038–8043 (2000).
Shi, Q., Thresher, R., Sancar, A. & Griffith, J. Electron microscopic study of (A)BC excinuclease—DNA is sharply bent in the UvrB-DNA complex. J. Mol. Biol. 226, 425–432 (1992).
Lin, J.J., Phillips, A.M., Hearst, J.E. & Sancar, A. Active site of (A)BC excinuclease: II. Binding, bending and catalysis mutants of UvrB reveal a direct role in 3′ and an indirect role in 5′ incision. J. Biol. Chem. 267, 17693–17700 (1992).
Verhoeven, E.E., Wyman, C., Moolenaar, G.F., Hoeijmakers, J.H. & Goosen, N. Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J. 20, 601–611 (2001).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Laskowski, R.A., McArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
This work has been supported by grants to C.K. from the NIH (GM 070873) and from the Pew Scholars Program in the Biomedical Sciences, and in part by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (B.V.H.). The National Synchrotron Light Source in Brookhaven is supported by the US Department of Energy and the NIH, and beamline X26C is supported in part by the State University of New York at Stony Brook and its Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
UvrB Y95A EMSA and incision data (PDF 580 kb)
Supplementary Table 1
Contacts between UvrB and the DNA (PDF 42 kb)
Rights and permissions
About this article
Cite this article
Truglio, J., Karakas, E., Rhau, B. et al. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol 13, 360–364 (2006). https://doi.org/10.1038/nsmb1072
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1072
This article is cited by
-
Global genome and transcription-coupled nucleotide excision repair pathway in prokaryotes
Journal of Biosciences (2023)
-
Single-molecule live-cell imaging visualizes parallel pathways of prokaryotic nucleotide excision repair
Nature Communications (2020)
-
Single-molecule imaging reveals molecular coupling between transcription and DNA repair machinery in live cells
Nature Communications (2020)
-
Homology Modeling, Molecular Docking and DNA Binding Studies of Nucleotide Excision Repair UvrC Protein from M. tuberculosis
The Protein Journal (2013)
-
Structure and mechanism of the UvrA–UvrB DNA damage sensor
Nature Structural & Molecular Biology (2012)