Abstract
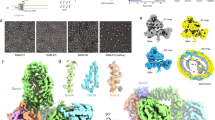
In Gram-negative bacteria, the assembly of β-barrel outer-membrane proteins (OMPs) requires the β-barrel–assembly machinery (BAM) complex. We determined the crystal structure of the 200-kDa BAM complex from Escherichia coli at 3.55-Å resolution. The structure revealed that the BAM complex assembles into a hat-like shape, in which the BamA β-barrel domain forms the hat's crown embedded in the outer membrane, and its five polypeptide transport–associated (POTRA) domains interact with the four lipoproteins BamB, BamC, BamD and BamE, thus forming the hat's brim in the periplasm. The assembly of the BAM complex creates a ring-like apparatus beneath the BamA β-barrel in the periplasm and a potential substrate-exit pore located at the outer membrane–periplasm interface. The complex structure suggests that the chaperone-bound OMP substrates may feed into the chamber of the ring-like apparatus and insert into the outer membrane via the potential substrate-exit pore in an energy-independent manner.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chacinska, A., Koehler, C.M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Walther, D.M., Rapaport, D. & Tommassen, J. Biogenesis of beta-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell. Mol. Life Sci. 66, 2789–2804 (2009).
Webb, C.T., Heinz, E. & Lithgow, T. Evolution of the β-barrel assembly machinery. Trends Microbiol. 20, 612–620 (2012).
Tommassen, J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156, 2587–2596 (2010).
Dalbey, R.E., Wang, P. & Kuhn, A. Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 (2011).
White, S.H. & von Heijne, G. How translocons select transmembrane helices. Annu. Rev. Biophys. 37, 23–42 (2008).
Osborne, A.R., Rapoport, T.A. & van den Berg, B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 21, 529–550 (2005).
Hagan, C.L., Silhavy, T.J. & Kahne, D. β-barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 (2011).
Pugsley, A.P. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57, 50–108 (1993).
Knowles, T.J., Scott-Tucker, A., Overduin, M. & Henderson, I.R. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206–214 (2009).
Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli . Cell 121, 235–245 (2005).
Ricci, D.P. & Silhavy, T.J. The Bam machine: a molecular cooper. Biochim. Biophys. Acta 1818, 1067–1084 (2012).
Rigel, N.W. & Silhavy, T.J. Making a beta-barrel: assembly of outer membrane proteins in Gram-negative bacteria. Curr. Opin. Microbiol. 15, 189–193 (2012).
Malinverni, J.C. et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli . Mol. Microbiol. 61, 151–164 (2006).
Roman-Hernandez, G., Peterson, J.H. & Bernstein, H.D. Reconstitution of bacterial autotransporter assembly using purified components. eLife 3, e04234 (2014).
Hagan, C.L., Kim, S. & Kahne, D. Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892 (2010).
Sánchez-Pulido, L., Devos, D., Genevrois, S., Vicente, M. & Valencia, A. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28, 523–526 (2003).
Jiang, J.H., Tong, J., Tan, K.S. & Gabriel, K. From evolution to pathogenesis: the link between β-barrel assembly machineries in the outer membrane of mitochondria and gram-negative bacteria. Int. J. Mol. Sci. 13, 8038–8050 (2012).
Voulhoux, R., Bos, M.P., Geurtsen, J., Mols, M. & Tommassen, J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 (2003).
Noinaj, N. et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 (2013).
Ni, D. et al. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli . FASEB J. 28, 2677–2685 (2014).
Albrecht, R. et al. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr. D Biol. Crystallogr. 70, 1779–1789 (2014).
Gruss, F. et al. The structural basis of autotransporter translocation by TamA. Nat. Struct. Mol. Biol. 20, 1318–1320 (2013).
Noinaj, N., Fairman, J.W. & Buchanan, S.K. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J. Mol. Biol. 407, 248–260 (2011).
Heuck, A., Schleiffer, A. & Clausen, T. Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J. Mol. Biol. 406, 659–666 (2011).
Kim, K.H., Aulakh, S. & Paetzel, M. Crystal structure of β-barrel assembly machinery BamCD protein complex. J. Biol. Chem. 286, 39116–39121 (2011).
Jansen, K.B., Baker, S.L. & Sousa, M.C. Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine. J. Biol. Chem. 290, 2126–2136 (2015).
Albrecht, R. & Zeth, K. Structural basis of outer membrane protein biogenesis in bacteria. J. Biol. Chem. 286, 27792–27803 (2011).
Sandoval, C.M., Baker, S.L., Jansen, K., Metzner, S.I. & Sousa, M.C. Crystal structure of BamD: an essential component of the β-barrel assembly machinery of gram-negative bacteria. J. Mol. Biol. 409, 348–357 (2011).
Dong, C., Hou, H.F., Yang, X., Shen, Y.Q. & Dong, Y.H. Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta Crystallogr. D Biol. Crystallogr. 68, 95–101 (2012).
Knowles, T.J. et al. Structure and function of BamE within the outer membrane and the β-barrel assembly machine. EMBO Rep. 12, 123–128 (2011).
Gatzeva-Topalova, P.Z., Walton, T.A. & Sousa, M.C. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16, 1873–1881 (2008).
Kim, S. et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 (2007).
Gatzeva-Topalova, P.Z., Warner, L.R., Pardi, A. & Sousa, M.C. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18, 1492–1501 (2010).
Knowles, T.J. et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 68, 1216–1227 (2008).
Zhang, H. et al. High-resolution structure of a new crystal form of BamA POTRA4-5 from Escherichia coli . Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 734–738 (2011).
Clantin, B. et al. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317, 957–961 (2007).
Stenberg, F. et al. Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280, 34409–34419 (2005).
Kim, K.H. & Paetzel, M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. J. Mol. Biol. 406, 667–678 (2011).
Ricci, D.P., Hagan, C.L., Kahne, D. & Silhavy, T.J. Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc. Natl. Acad. Sci. USA 109, 3487–3491 (2012).
Bos, M.P., Robert, V. & Tommassen, J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 8, 1149–1154 (2007).
Gessmann, D. et al. Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc. Natl. Acad. Sci. USA 111, 5878–5883 (2014).
Bakelar, J., Buchanan, S.K. & Noinaj, N. The structure of the β-barrel assembly machinery complex. Science 351, 180–186 (2016).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
We are grateful to T. Silhavy (Princeton University) for generously providing the E. coli strain JCM320. We also thank X. Zhang and H. Wu for valuable discussions; R. Zhang and Shanghai Synchrotron Radiation Facility beamline scientists for scheduling beamline time; F. Yang, L. Niu, M. Zhang, L. Shu, Z. Xie and X. Ding from the Mass Spectrometry Core Facility of the Institute of Biophysics, Chinese Academy of Sciences, for help with mass spectrometry analysis of the BAM complex; and the National Supercomputing Center Tianjin Center (Tianhe), China, for computational resources. This work was supported by grants from the Ministry of Science and Technology (2012CB917302 and 2013CB910603 to Y.H.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB080202 to Y.H.) and the National Natural Science Foundation of China (31170698 and 31470743 to Y.H.).
Author information
Authors and Affiliations
Contributions
Y.H. supervised the project. L.H., J.Z., Y.W., X.Y., H.Z. and D.N. performed protein purification, crystallization and diffraction data collection. Y.H. and B.C. determined the structure and built the model. Y.H. wrote the manuscript. All authors contributed to data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
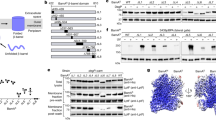
Supplementary Figure 1 Schematic domain organization of the BAM complex and characterization of the purified BAM complex.
a. Schematic domain structures of the mature proteins of BamA, BamB, BamC, BamD and BamE. b. Size exclusion chromatographic profile of the purified BAM complex from bacterial OMs on Superose 6 10/300 GL column. c. 15% SDS-PAGE analysis of fractions from the elution peak (left) and of the BAM complex crystals (dissolved from 50 crystals) to show that BamC remained intact in the crystals (right).
Supplementary Figure 2 Overlay of isolated POTRA domains or β-barrel–attached POTRA domains of BamA.
a. Overlay of POTRA4 domains from BamA in the BAM complex (BAM-POTRA1-5), POTRA4-5 (PDB: 3Q6B), POTRA1-4 (PDBs: 3EFC and 2QCZ) to show the remarkably different inter-POTRA interfaces and orientations. b. Structural overlay of the β-barrel domains of BamA from the BAM complex (BAM-BamA), BamA lacking POTRA1-4 domains (BamA_POTRA5, PDB: 4C4V) and HdBamA lacking POTRA1-3 domains (HdBamA_POTRA4-5, PDB: 4K3C) to show the remarkably different interdomain orientation between BamA β barrel and the POTRA5 domain.
Supplementary Figure 3 Strand β1 of the BamA β-barrel might participate in β-augmentation.
Residues T243, S324, D399 (or T400) and G429 of BamA are located at the middle point of a β strand of POTRA3, POTRA4, POTRA5 and strand β1 of BamA β barrel, respectively. These β strands are located within the chamber of the BAM complex structure and have a potential role in participating β-augmentation for OMP substrates. a. G429P mutant was lethal to the bacteria while other mutants were not. b. G429P mutant was lethal in the presence of 10 μM IPTG. c. Western blot showing that G429P mutant protein was expressed in a similar level to that of the wild-type in the cell. d. Purified G429P mutant protein was well-folded. Purified G429P protein from membranes exhibited apparently heat-modifiable mobility on a 15% SDS-PAGE gel.
Supplementary Figure 4 Structural comparisons of BamE and BamCD complexes.
a. Overlay of BamE in the BAM complex with the isolated BamE structures from crystal structure (BamE-X-ray, PDB: 2YH9) and NMR structure (BamE-NMR, PDB: 2KXX). Overlay of the three β strands of BamE structures reveals an RMSD of 2.5 Å (35 aligned Cα atom pairs) and of 2.2 Å (35 aligned Cα atom pairs) between BamE in the BAM complex and BamE crystal structure or BamE NMR structure. b. Structural comparison of BamCD complex with that in the BAM complex. Neither the BamC_N domain nor the BamC_C domain was visible in the BAM complex structure.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 665 kb)
Supplementary Data Set 1
Uncropped images of western blots for Figure 2d, Figure 4e and Figure 4f (PDF 647 kb)
Rights and permissions
About this article
Cite this article
Han, L., Zheng, J., Wang, Y. et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23, 192–196 (2016). https://doi.org/10.1038/nsmb.3181
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3181
This article is cited by
-
Escherichia coli displays a conserved membrane proteomic response to a range of alcohols
Biotechnology for Biofuels and Bioproducts (2023)
-
Surveying membrane landscapes: a new look at the bacterial cell surface
Nature Reviews Microbiology (2023)
-
Peptidoglycan maturation controls outer membrane protein assembly
Nature (2022)
-
Burkholderia cenocepacia transcriptome during the early contacts with giant plasma membrane vesicles derived from live bronchial epithelial cells
Scientific Reports (2021)
-
The role of membrane destabilisation and protein dynamics in BAM catalysed OMP folding
Nature Communications (2021)