Abstract
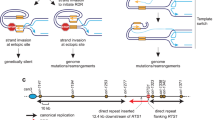
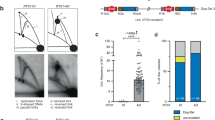
To maintain genetic stability, DNA must be replicated only once per cell cycle, and replication must be completed even when individual replication forks are inactivated. Because fork inactivation is common, passive convergence of an adjacent fork is insufficient to rescue all inactive forks. Thus, eukaryotic cells have evolved homologous recombination–dependent mechanisms to restart persistent inactive forks. Completing DNA synthesis via homologous recombination–restarted replication (HoRReR) ensures cell survival, but at a cost. One such cost is increased mutagenesis because HoRReR is more error prone than canonical replication. This increased error rate implies the HoRReR mechanism is distinct from that of a canonical fork. Here we demonstrate, in Schizosaccharomyces pombe, that a DNA sequence duplicated by HoRReR during S phase is replicated semiconservatively, but both the leading and lagging strands are synthesized by DNA polymerase δ.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lambert, S. & Carr, A.M. Replication stress and genome rearrangements: lessons from yeast models. Curr. Opin. Genet. Dev. 23, 132–139 (2013).
Liu, P., Carvalho, C.M., Hastings, P.J. & Lupski, J.R. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 22, 211–220 (2012).
Sun, Z. et al. Replicative mechanisms of CNV formation preferentially occur as intrachromosomal events: evidence from Potocki-Lupski duplication syndrome. Hum. Mol. Genet. 22, 749–756 (2013).
Carvalho, C.M. et al. Replicative mechanisms for CNV formation are error prone. Nat. Genet. 45, 1319–1326 (2013).
Bignell, G.R. et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 17, 1296–1303 (2007).
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007).
Barlow, J.H. et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 152, 620–632 (2013).
Lambert, S. & Carr, A.M. Impediments to replication fork movement: stabilisation, reactivation and genome instability. Chromosoma 122, 33–45 (2013).
Errico, A. & Costanzo, V. Mechanisms of replication fork protection: a safeguard for genome stability. Crit. Rev. Biochem. Mol. Biol. 47, 222–235 (2012).
Lopes, M. et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412, 557–561 (2001).
Higgs, M.R. et al. BOD1L is required to suppress deleterious resection of stressed replication forks. Mol. Cell 59, 462–477 (2015).
Ge, X.Q., Jackson, D.A. & Blow, J.J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 21, 3331–3341 (2007).
Lambert, S., Watson, A., Sheedy, D.M., Martin, B. & Carr, A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121, 689–702 (2005).
Anand, R.P., Lovett, S.T. & Haber, J.E. Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 5, a010397 (2013).
Jain, S. et al. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 23, 291–303 (2009).
Malkova, A. & Ira, G. Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 23, 271–279 (2013).
Lambert, S. et al. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell 39, 346–359 (2010).
Mohebi, S., Mizuno, K., Watson, A., Carr, A.M. & Murray, J.M. Checkpoints are blind to replication restart and recombination intermediates that result in gross chromosomal rearrangements. Nat. Commun. 6, 6357 (2015).
Nguyen, M.O., Jalan, M., Morrow, C.A., Osman, F. & Whitby, M.C. Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. eLife 4, e04539 (2015).
Mizuno, K., Lambert, S., Baldacci, G., Murray, J.M. & Carr, A.M. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 23, 2876–2886 (2009).
Smith, C.E., Llorente, B. & Symington, L.S. Template switching during break-induced replication. Nature 447, 102–105 (2007).
Deem, A. et al. Break-induced replication is highly inaccurate. PLoS Biol. 9, e1000594 (2011).
Costantino, L. et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343, 88–91 (2014).
Iraqui, I. et al. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 8, e1002976 (2012).
Mizuno, K., Miyabe, I., Schalbetter, S.A., Carr, A.M. & Murray, J.M. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493, 246–249 (2013).
Eydmann, T. et al. Rtf1-mediated eukaryotic site-specific replication termination. Genetics 180, 27–39 (2008).
Calzada, A., Hodgson, B., Kanemaki, M., Bueno, A. & Labib, K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19, 1905–1919 (2005).
Miyabe, I., Kunkel, T.A. & Carr, A.M. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 7, e1002407 (2011).
Daigaku, Y. et al. A global profile of replicative polymerase usage. Nat. Struct. Mol. Biol. 22, 192–198 (2015).
Kai, M. & Wang, T.S. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 17, 64–76 (2003).
Donnianni, R.A. & Symington, L.S. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl. Acad. Sci. USA 110, 13475–13480 (2013).
Saini, N. et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502, 389–392 (2013).
Wilson, M.A. et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–396 (2013).
Meselson, M. & Stahl, F.W. The replication of DNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 44, 671–682 (1958).
Kesti, T., Flick, K., Keranen, S., Syvaoja, J.E. & Wittenberg, C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3, 679–685 (1999).
Waga, S. & Stillman, B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369, 207–212 (1994).
Handa, T., Kanke, M., Takahashi, T.S., Nakagawa, T. & Masukata, H. DNA polymerization-independent functions of DNA polymerase epsilon in assembly and progression of the replisome in fission yeast. Mol. Biol. Cell 23, 3240–3253 (2012).
Schlacher, K., Wu, H. & Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012).
Petermann, E., Orta, M.L., Issaeva, N., Schultz, N. & Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 (2010).
Carr, A.M. & Lambert, S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J. Mol. Biol. 425, 4733–4744 (2013).
Kunkel, T.A. & Burgers, P.M. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 (2008).
Georgescu, R.E. et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat. Struct. Mol. Biol. 21, 664–670 (2014).
Reijns, M.A. et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518, 502–506 (2015).
Clausen, A.R. et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 22, 185–191 (2015).
Lujan, S.A. et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 24, 1751–1764 (2014).
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005).
Gorgoulis, V.G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005).
Bartkova, J. et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 (2006).
Bester, A.C. et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 (2011).
Dominguez-Sola, D. et al. Non-transcriptional control of DNA replication by c-Myc. Nature 448, 445–451 (2007).
Jones, R.M. et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene 32, 3744–3753 (2013).
Di Micco, R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 (2006).
Neelsen, K.J., Zanini, I.M., Herrador, R. & Lopes, M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol. 200, 699–708 (2013).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 (1991).
Watson, A.T., Garcia, V., Bone, N., Carr, A.M. & Armstrong, J. Gene tagging and gene replacement using recombinase-mediated cassette exchange in Schizosaccharomyces pombe. Gene 407, 63–74 (2008).
Lea, D.E. & Coulson, C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49, 264–285 (1949).
Watson, A.T. et al. Optimisation of the Schizosaccharomyces pombe urg1 expression system. PLoS ONE 8, e83800 (2013).
Arcangioli, B. Fate of mat1 DNA strands during mating-type switching in fission yeast. EMBO Rep. 1, 145–150 (2000).
Acknowledgements
This work was supported by Project Z01 ES065070 (T.A.K.) from the Division of Intramural Research of the US National Institutes of Health, by Medical Research Council (UK) grants G0801078 and G1100074 (A.M.C.) and by European Research Council grant 268788-SMI-DDR (A.M.C.). We thank H. Masukata (Osaka University) for the cdc20::hphMX6-Pnmt1-cdc20CTD strain.
Author information
Authors and Affiliations
Contributions
A.M.C. conceived the study. A.M.C., T.A.K., I.M. and J.M.M. designed the experimental approach. I.M., K'I.M., S.M., Y.D., A.K. and M.S. performed experiments and interpreted data. I.M. and A.M.C. wrote the manuscript. T.A.K. and J.M.M. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data Set 1
Full autoradiographs of all blots used in the figures (PDF 13572 kb)
Rights and permissions
About this article
Cite this article
Miyabe, I., Mizuno, K., Keszthelyi, A. et al. Polymerase δ replicates both strands after homologous recombination–dependent fork restart. Nat Struct Mol Biol 22, 932–938 (2015). https://doi.org/10.1038/nsmb.3100
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3100
This article is cited by
-
Gene duplication and deletion caused by over-replication at a fork barrier
Nature Communications (2023)
-
Global landscape of replicative DNA polymerase usage in the human genome
Nature Communications (2022)
-
How asymmetric DNA replication achieves symmetrical fidelity
Nature Structural & Molecular Biology (2021)
-
Replication dynamics of recombination-dependent replication forks
Nature Communications (2021)
-
The nuclear pore primes recombination-dependent DNA synthesis at arrested forks by promoting SUMO removal
Nature Communications (2020)