Abstract
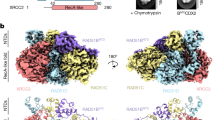
Mutations in BRCA2 increase susceptibility to breast, ovarian and prostate cancers. The product of human BRCA2, BRCA2 protein, has a key role in the repair of DNA double-strand breaks and interstrand cross-links by RAD51-mediated homologous recombination. Here, we present a biochemical and structural characterization of full-length (3,418 amino acid) BRCA2, alone and in complex with RAD51. We show that BRCA2 facilitates nucleation of RAD51 filaments at multiple sites on single-stranded DNA. Three-dimensional EM reconstructions revealed that BRCA2 exists as a dimer and that two oppositely oriented sets of RAD51 molecules bind the dimer. Single-stranded DNA binds along the long axis of BRCA2, such that only one set of RAD51 monomers can form a productive complex with DNA and establish filament formation. Our data define the molecular mechanism by which this tumor suppressor facilitates RAD51-mediated homologous-recombinational repair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Stratton, M.R. & Rahman, N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 40, 17–22 (2008).
Wooster, R. et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 378, 789–792 (1995).
King, M.C., Marks, J.H., Mandell, J.B. & Grp, N.Y.B.C.S. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646 (2003).
Lindahl, T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993).
Jackson, S.P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Venkitaraman, A.R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108, 171–182 (2002).
Roy, R., Chun, J. & Powell, S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68–78 (2012).
Moynahan, M.E., Pierce, A.J. & Jasin, M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 (2001).
Lomonosov, M., Anand, S., Sangrithi, M., Davies, R. & Venkitaraman, A.R. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 17, 3017–3022 (2003).
Schlacher, K. et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 (2011).
Yuan, S.-S.F. et al. BRCA2 is required for ionizing radiation-induced assembly of RAD51 complex in vivo. Cancer Res. 59, 3547–3551 (1999).
Tarsounas, M., Davies, D. & West, S.C. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 22, 1115–1123 (2003).
Bork, P., Blomberg, N. & Nilges, M. Internal repeats in the BRCA2 protein sequence. Nat. Genet. 13, 22–23 (1996).
Bignell, G., Micklem, G., Stratton, M.R., Ashworth, A. & Wooster, R. The BRC repeats are conserved in mammalian BRCA2 proteins. Hum. Mol. Genet. 6, 53–58 (1997).
Davies, A.A. et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7, 273–282 (2001).
Esashi, F., Galkin, V.E., Yu, X., Egelman, E.H. & West, S.C. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 14, 468–474 (2007).
Carreira, A. et al. The BRC repeats of BRCA2 modulate the DNA-binding specificity of RAD51. Cell 136, 1032–1043 (2009).
Pellegrini, L. et al. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 420, 287–293 (2002).
Tal, A., Arbel-Goren, R. & Stavans, J. Cancer-associated mutations in BRC domains of BRCA2 affect homologous recombination induced by RAD51. J. Mol. Biol. 393, 1007–1012 (2009).
Thorslund, T. et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1263–1265 (2010).
Jensen, R.B., Carreira, A. & Kowalczykowski, S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 (2010).
Liu, J., Doty, T., Gibson, B. & Heyer, W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 (2010).
Lee, S.A., Roques, C., Magwood, A.C., Masson, J.Y. & Baker, M.D. Recovery of deficient homologous recombination in BRCA2-depleted mouse cells by wild-type RAD51 expression. DNA Repair (Amst.) 8, 170–181 (2009).
Buisson, R. et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17, 1247–1254 (2010).
Dray, E. et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 17, 1255–1259 (2010).
Rahman, N. et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 39, 165–167 (2007).
Sy, S.M.H., Huen, M.S.Y., Zhu, Y.Y. & Chen, J.J. PALB2 regulates recombinational repair through chromatin association and oligomerization. J. Biol. Chem. 284, 18302–18310 (2009).
Thorslund, T., Esashi, F. & West, S.C. Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 26, 2915–2922 (2007).
Yang, H. et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297, 1837–1848 (2002).
van Heel, M. et al. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 (2000).
van Heel, M., Harauz, G., Orlova, E.V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Rosenthal, P.B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Sauerwald, A. et al. Structure of active dimeric human telomerase. Nat. Struct. Mol. Biol. 20, 454–460 (2013).
Wong, A.K.C., Pero, R., Ormonde, P.A., Tavtigian, S.V. & Bartel, P.L. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene BRCA2. J. Biol. Chem. 272, 31941–31944 (1997).
Goddard, T.D., Huang, C.C. & Ferrin, T.E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007).
Benson, F.E., Stasiak, A. & West, S.C. Purification and characterisation of the human RAD51 protein, an analogue of E. coli RecA. EMBO J. 13, 5764–5771 (1994).
Baumann, P., Benson, F.E. & West, S.C. Human RAD51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87, 757–766 (1996).
Shivji, M.K.K. et al. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc. Natl. Acad. Sci. USA 106, 13254–13259 (2009).
Ho, Y. et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 (2002).
Sung, P. & Robberson, D.L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82, 453–461 (1995).
Baumann, P. & West, S.C. The human RAD51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 16, 5198–5206 (1997).
Hilario, J., Amitani, I., Baskin, R.J. & Kowalczykowski, S.C. Direct imaging of human RAD51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. USA 106, 361–368 (2009).
van der Heijden, T. et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 35, 5646–5657 (2007).
Zhou, Q. et al. Dss1 interaction with Brh2 as a regulatory mechanism for recombinational repair. Mol. Cell. Biol. 27, 2512–2526 (2007).
Jeyasekharan, A.D. et al. DNA damage regulates the mobility of BRCA2 within the nucleoplasm of living cells. Proc. Natl. Acad. Sci. USA 107, 21937–21942 (2010).
Bakkenist, C.J. & Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 (2003).
Saeki, H. et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl. Acad. Sci. USA 103, 8768–8773 (2006).
Bochkarev, A. & Bochkareva, E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 14, 36–42 (2004).
Fan, J. & Pavletich, N.P. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 (2012).
Bianchi, M., Das Gupta, C. & Radding, C.M. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell 34, 931–939 (1983).
Bortner, C. & Griffith, J. Three-stranded paranemic joints: architecture, topological constraints and movement. J. Mol. Biol. 215, 623–634 (1990).
McIlwraith, M.J. et al. Reconstitution of the strand invasion step of double-strand break repair using human RAD51, RAD52 and RPA proteins. J. Mol. Biol. 304, 151–164 (2000).
Namsaraev, E.A. & Berg, P. Rad51 uses one mechanism to drive DNA strand exchange in both directions. J. Biol. Chem. 275, 3970–3976 (2000).
Yang, H., Li, Q., Holloman, W.K. & Pavletich, N.P. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433, 653–657 (2005).
West, S.C., Cassuto, E. & Howard-Flanders, P. Heteroduplex formation by RecA protein: polarity of strand exchange. Proc. Natl. Acad. Sci. USA 78, 6149–6153 (1981).
Kahn, R., Cunningham, R.P., DasGupta, C. & Radding, C.M. Polarity of heteroduplex formation promoted by Escherichia coli RecA protein. Proc. Natl. Acad. Sci. USA 78, 4786–4790 (1981).
Cox, M.M. & Lehman, I.R. Directionality and polarity in RecA protein-promoted branch migration. Proc. Natl. Acad. Sci. USA 78, 6018–6022 (1981).
Baumann, P., Benson, F.E., Hajibagheri, N. & West, S.C. Purification of human RAD51 protein by selective spermidine precipitation. Mutat. Res. DNA Repair 384, 65–72 (1997).
Rosenthal, P.B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Henderson, R. et al. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J. Mol. Biol. 413, 1028–1046 (2011).
Sauerwald, A. et al. Structure of active dimeric human telomerase. Nat. Struct. Mol. Biol. 20, 454–460 (2013).
Wall, J.S., Hainfeld, J.F. & Simon, M.N. Scanning transmission electron microscopy of nuclear structures. Methods Cell Biol. 53, 139–164 (1998).
Acknowledgements
We thank R. Carzaniga and L. Collinson at the London Research Institute Electron Microscope facility for advice and input and our colleagues for their comments. We thank Innova Biosciences for providing the InnovaCoat conjugation kit. This work was supported by the UK Medical Research Council and the Wellcome Trust (X.Z.) and by Cancer Research UK, the European Research Council, the Breast Cancer Campaign, Swiss Bridge and the Louis-Jeantet foundation (S.C.W.). L.M. was supported in part by an European Molecular Biology Organization Fellowship. J.S. was supported in part by a Marie Curie Intra-European Fellowship (MC-IEF-625826).
Author information
Authors and Affiliations
Contributions
S.C.W. and X.Z. conceived the original ideas for this study. All authors contributed to experimental designs. T.S., J.S., E.H.K. and M.J.M. performed the experiments. L.M. and T.P. contributed to initial characterizations of the BRCA2 protein. T.S., J.S., E.H.K., M.J.M., S.C.W. and X.Z. analyzed the data. X.Z. and S.C.W. wrote the main text, and all authors contributed to the writing of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 BRCA2 and BRCA2–RAD51 purification and binding to DNA substrates.
(a) SDS-PAGE showing the purified proteins used for the electron microscopy studies. (b) BRCA2 stimulates RAD51-ssDNA nucleoprotein filament formation, as determined by gel-shift assays. (c) Binding of BRCA2 to gapped DNA, as determined by gel-shift assays.
Supplementary Figure 2 Electron micrographs of BRCA2 and BRCA2–RAD51 complex.
(a) Micrograph and representative selected individual particles of BRCA2. (b) Micrograph and representative selected individual particles of BRCA2-RAD51. Scale bar is 100 nm.
Supplementary Figure 3 Image processing of BRCA2 reconstruction and antibody labeling.
(a) Initial eigen-images show a clear 2-fold component in the particles. (b) Non-symmetrized 3D reconstruction in orthogonal reprojections (top) and slices though the 3D (bottom) showing 2-fold component (c) Representative class averages (CA) and reprojections from the final 3D reconstruction. (d) Locations of antibody against Flag tag (blue) and BRC repeats (magenta) through triangulation for top and side views.
Supplementary Figure 4 Resolution of EM reconstructions and tilt validation.
(a) Resolution as determined by Fourier shell correlation coefficient using 0.5, ½-bit and 0.143 criteria for binary masking (bm), soft spherical masking (csm) or no masking (nm) applied to the BRCA2 and BRCA2-RAD51 reconstructions. On the right are the corresponding Euler angle distributions of classums that gave rise to the final 3D reconstructions. (b) Particles every 10º from a 0-40º tilt-series with 5º steps and matching projections of BRCA2 (left) and BRCA2-RAD51 (right), and (c) tables of nominal versus measured tilts for BRCA2 (left) and BRCA2-RAD51 (right).
Supplementary Figure 5 Image processing of BRCA2–RAD51 reconstruction.
(a) Initial eigenimages show a clear 2-fold component in the particles. (b) Non-symmetrized 3D reconstruction in orthogonal reprojections (top) and slices though the 3D (bottom) showing 2-fold component (c) Representative class averages (CA) and reprojections from the 3D reconstruction.
Supplementary Figure 6 RecA and RAD51 filament orientations.
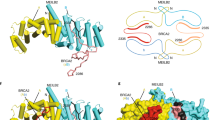
(a) In addition to the conserved ATPase core domain, RecA and RAD51 have distinct additional domains positioned on opposite sides of the ATPase core relative to ssDNA. (b) Schematic diagram of how RecA (yellow and red) and RAD51 (yellow and blue) subunits may be added to the growing filament. The ATPase domain is represented in yellow. The respective loops that only become ordered and stabilized in the filament are shown as broken (disordered) and solid lines (ordered). (c) RecA-ssDNA filament as observed in the crystal structure and RAD51 filament modelled onto the RecA filament. The unique domains are boxed.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 4135 kb)
Rights and permissions
About this article
Cite this article
Shahid, T., Soroka, J., Kong, E. et al. Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor. Nat Struct Mol Biol 21, 962–968 (2014). https://doi.org/10.1038/nsmb.2899
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2899
This article is cited by
-
Significant association of BRCA1 (rs1799950), BRCA2 (rs144848) and TP53 (rs1042522) polymorphism with breast cancer risk in Pashtun population of Khyber Pakhtunkhwa, Pakistan
Molecular Biology Reports (2023)
-
A novel somatic BRCA2 point mutation in a metastatic pancreatic cancer patient: a case report
BMC Medical Genomics (2021)
-
Nucleotide proofreading functions by nematode RAD51 paralogs facilitate optimal RAD51 filament function
Nature Communications (2021)
-
Structure of a meiosis-specific complex central to BRCA2 localization at recombination sites
Nature Structural & Molecular Biology (2021)
-
Fanconi anemia pathway and its relationship with cancer
Genome Instability & Disease (2021)