Abstract
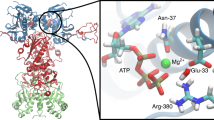
The 70-kilodalton (kDa) heat-shock proteins (Hsp70s) are ubiquitous molecular chaperones essential for cellular protein folding and proteostasis. Each Hsp70 has two functional domains: a nucleotide-binding domain (NBD), which binds and hydrolyzes ATP, and a substrate-binding domain (SBD), which binds extended polypeptides. NBD and SBD interact little when in the presence of ADP; however, ATP binding allosterically couples the polypeptide- and ATP-binding sites. ATP binding promotes polypeptide release; polypeptide rebinding stimulates ATP hydrolysis. This allosteric coupling is poorly understood. Here we present the crystal structure of an intact ATP-bound Hsp70 from Escherichia coli at 1.96-Å resolution. The ATP-bound NBD adopts a unique conformation, forming extensive interfaces with an SBD that has changed radically, having its α-helical lid displaced and the polypeptide-binding channel of its β-subdomain restructured. These conformational changes, together with our biochemical assays, provide a structural explanation for allosteric coupling in Hsp70 activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hartl, F.U., Bracher, A. & Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011).
Balch, W.E., Morimoto, R.I., Dillin, A. & Kelly, J.W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008).
Bukau, B. & Horwich, A.L. The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366 (1998).
Mayer, M.P. Gymnastics of molecular chaperones. Mol. Cell 39, 321–331 (2010).
Zuiderweg, E.R. et al. Allostery in the Hsp70 Chaperone Proteins. Top. Curr. Chem. 328, 99–153 (2013).
Rüdiger, S., Buchberger, A. & Bukau, B. Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4, 342–349 (1997).
Flaherty, K.M., DeLuca-Flaherty, C. & McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346, 623–628 (1990).
Sriram, M., Osipiuk, J., Freeman, B., Morimoto, R. & Joachimiak, A. Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 5, 403–414 (1997).
Harrison, C.J., Hayer-Hartl, M., Di Liberto, M., Hartl, F. & Kuriyan, J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276, 431–435 (1997).
Jiang, J., Prasad, K., Lafer, E.M. & Sousa, R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol. Cell 20, 513–524 (2005).
Chang, Y.W., Sun, Y.J., Wang, C. & Hsiao, C.D. Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. J. Biol. Chem. 283, 15502–15511 (2008).
Zhu, X. et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 (1996).
Wang, H. et al. NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction. Biochemistry 37, 7929–7940 (1998).
Pellecchia, M. et al. Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Struct. Biol. 7, 298–303 (2000).
Liebscher, M. & Roujeinikova, A. Allosteric coupling between the lid and interdomain linker in DnaK revealed by inhibitor binding studies. J. Bacteriol. 191, 1456–1462 (2009).
Buchberger, A. et al. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J. Biol. Chem. 270, 16903–16910 (1995).
Swain, J.F. et al. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 (2007).
Bertelsen, E.B., Chang, L., Gestwicki, J.E. & Zuiderweg, E.R. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. USA 106, 8471–8476 (2009).
Schmid, D., Baici, A., Gehring, H. & Christen, P. Kinetics of molecular chaperone action. Science 263, 971–973 (1994).
Flynn, G.C., Chappell, T.G. & Rothman, J.E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245, 385–390 (1989).
Flaherty, K.M., Wilbanks, S.M., DeLuca-Flaherty, C. & McKay, D.B. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment. J. Biol. Chem. 269, 12899–12907 (1994).
Jiang, J. et al. Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28, 422–433 (2007).
Liu, Q. & Hendrickson, W.A. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120 (2007).
Smock, R.G. et al. An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol. Syst. Biol. 6, 414 (2010).
Mapa, K. et al. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol. Cell 38, 89–100 (2010).
Marcinowski, M. et al. Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 (2011).
Schlecht, R., Erbse, A.H., Bukau, B. & Mayer, M.P. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 (2011).
McCarty, J.S. & Walker, G.C. DnaK as a thermometer: threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc. Natl. Acad. Sci. USA 88, 9513–9517 (1991).
Osipiuk, J., Georgopoulos, C. & Zylicz, M. Initiation of lambda DNA replication. The Escherichia coli small heat shock proteins, DnaJ and GrpE, increase DnaK's affinity for the lambda P protein. J. Biol. Chem. 268, 4821–4827 (1993).
Xu, X. et al. The unique peptide substrate binding properties of 110 KDA heatshock protein (HSP110) determines its distinct chaperone activity. J. Biol. Chem. 287, 5661–5672 (2012).
Smock, R.G., Blackburn, M.E. & Gierasch, L.M. Conserved, disordered C terminus of DnaK enhances cellular survival upon stress and DnaK in vitro chaperone activity. J. Biol. Chem. 286, 31821–31829 (2011).
Liu, Q. et al. Structures from anomalous diffraction of native biological macromolecules. Science 336, 1033–1037 (2012).
Swain, J.F. & Gierasch, L.M. The changing landscape of protein allostery. Curr. Opin. Struct. Biol. 16, 102–108 (2006).
Vogel, M., Mayer, M.P. & Bukau, B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J. Biol. Chem. 281, 38705–38711 (2006).
Zhuravleva, A. & Gierasch, L.M. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc. Natl. Acad. Sci. USA 108, 6987–6992 (2011).
Kumar, D.P. et al. The four hydrophobic residues on the Hsp70 inter-domain linker have two distinct roles. J. Mol. Biol. 411, 1099–1113 (2011).
Bhattacharya, A. et al. Allostery in Hsp70 chaperones is transduced by subdomain rotations. J. Mol. Biol. 388, 475–490 (2009).
Rist, W., Graf, C., Bukau, B. & Mayer, M.P. Amide hydrogen exchange reveals conformational changes in hsp70 chaperones important for allosteric regulation. J. Biol. Chem. 281, 16493–16501 (2006).
Swain, J.F., Schulz, E.G. & Gierasch, L.M. Direct comparison of a stable isolated Hsp70 substrate-binding domain in the empty and substrate-bound states. J. Biol. Chem. 281, 1605–1611 (2006).
Lovell, S.C. et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50, 437–450 (2003).
Montgomery, D.L., Morimoto, R.I. & Gierasch, L.M. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J. Mol. Biol. 286, 915–932 (1999).
Gässler, C.S. et al. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. USA 95, 15229–15234 (1998).
Vogel, M., Bukau, B. & Mayer, M.P. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol. Cell 21, 359–367 (2006).
Davis, J.E., Voisine, C. & Craig, E.A. Intragenic suppressors of Hsp70 mutants: interplay between the ATPase- and peptide-binding domains. Proc. Natl. Acad. Sci. USA 96, 9269–9276 (1999).
Polier, S., Dragovic, Z., Hartl, F.U. & Bracher, A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079 (2008).
Zhuravleva, A., Clerico, E.M. & Gierasch, L.M. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 (2012).
Kityk, R., Kopp, J., Sinning, I. & Mayer, M.P. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 (2012).
Liu, Q., Liu, Q. & Hendrickson, W.A. Robust structural analysis of native biological macromolecules from multi-crystal anomalous diffraction data. Acta Crystallogr. D Biol. Crystallogr. (in the press).
Mossessova, E. & Lima, C.D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000).
Burkholder, W.F. et al. Mutations in the C-terminal fragment of DnaK affecting peptide binding. Proc. Natl. Acad. Sci. USA 93, 10632–10637 (1996).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 (2011).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
Read, R.J. & McCoy, A.J. Using SAD data in Phaser. Acta Crystallogr. D Biol. Crystallogr. 67, 338–344 (2011).
Cowtan, K.D. & Zhang, K.Y.J. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 72, 245–270 (1999).
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Adams, P.D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Laskowski, R.A., Macarthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Becker, J., Walter, W., Yan, W. & Craig, E.A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell Biol. 16, 4378–4386 (1996).
Lindahl, E., Hess, B. & van der Spoel, D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7, 306–317 (2001).
Balsera, M.A., Wriggers, W., Oono, Y. & Schulten, K. Principal component analysis and long time protein dynamics. J. Phys. Chem. 100, 2567–2572 (1996).
Ichiye, T. & Karplus, M. Collective motions in proteins: a covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins 11, 205–217 (1991).
Acknowledgements
We thank E. Craig, D. Logothetis, G. Tseng, L. Avery, L. Greene, J. Rife and C. Fox for critically reading the manuscript and providing insightful suggestions. We are grateful to J. Schwanof, R. Abramowitz and X. Yang (Brookhaven National Laboratory Beamline X4A and X4C) for their assistance in collecting diffraction data. We thank D. Kumar for technical support and C. Escalante for the PC1 photon counting spectrofluorimeter. This work was supported by startup funds from the Virginia Commonwealth University School of Medicine (to Qinglian Liu), a New Scholar Award in Aging from the Ellison Medical Foundation (AG-NS-0587-09 to Qinglian Liu) and a Grant-In-Aid Award from the American Heart Association (11GRNT7460003 to Qinglian Liu).
Author information
Authors and Affiliations
Contributions
Qinglian Liu designed the study and experiments. Qinglian Liu and L.Z. screened, identified and carried out the cloning for the crystallization construct of full-length DnaK. R.Q. and E.B.S. prepared the crystals for full-length DnaK and isolated SBD, respectively. Qun Liu and Qinglian Liu performed the structure determination and modeling. R.Q., E.B.S., K.Q.L., X.X., H.X., J.Y., J.L.W. and C.V. carried out the cloning of the mutations for biochemical analysis, protein purification, biochemical and genetic analysis. L.Z. performed computational analysis on the structures. Qinglian Liu, W.A.H. and L.Z. analyzed the structures, carried out data analysis and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1 and 2 (PDF 6412 kb)
Rights and permissions
About this article
Cite this article
Qi, R., Sarbeng, E., Liu, Q. et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol 20, 900–907 (2013). https://doi.org/10.1038/nsmb.2583
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2583
This article is cited by
-
A first-in-class inhibitor of Hsp110 molecular chaperones of pathogenic fungi
Nature Communications (2023)
-
The Hsc70 disaggregation machinery removes monomer units directly from α-synuclein fibril ends
Nature Communications (2021)
-
Conformational dynamics of free and membrane-bound human Hsp70 in model cytosolic and endo-lysosomal environments
Communications Biology (2021)
-
Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein
Nature Communications (2020)
-
Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response
Nature Communications (2020)