Abstract
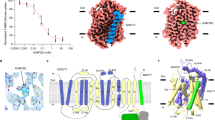
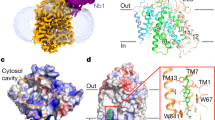
Here we present two structures of the major facilitator (MFS) xylose transporter XylE from Escherichia coli in inward open and partially occluded inward open conformations. These structures provide key information about the transport cycle of XylE and the closely related human GLUT transporters. This is, to our knowledge, the first MFS transporter structure determined in more than one conformational state, which may establish XylE as an important MFS model protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Augustin, R. IUBMB Life 62, 315–333 (2010).
Davis, E.O. & Henderson, P.J. J. Biol. Chem. 262, 13928–13932 (1987).
Newstead, S. et al. EMBO J. 30, 417–426 (2011).
Jardetzky, O. Nature 211, 969–970 (1966).
Sun, L. et al. Nature 490, 361–366 (2012).
Abramson, J. et al. Science 301, 610–615 (2003).
Huang, Y. et al. Science 301, 616–620 (2003).
Solcan, N. et al. EMBO J. 31, 3411–3421 (2012).
Carruthers, A. et al. Am. J. Physiol. Endocrinol. Metab. 297, E836–E848 (2009).
Law, C.J. et al. Annu. Rev. Microbiol. 62, 289–305 (2008).
Woestenenk, E.A., Hammarstrom, M., van den Berg, S., Hard, T. & Berglund, H. J. Struct. Funct. Genomics 5, 217–229 (2004).
Kabsch, W. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Strong, M. et al. Proc. Natl. Acad. Sci. USA 103, 8060–8065 (2006).
McCoy, A.J. et al. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Waterhouse, A.M., Procter, J.B., Martin, D.M., Clamp, M. & Barton, G.J. Bioinformatics 25, 1189–1191 (2009).
Acknowledgements
E.M.Q. was supported by The Danish Council for Independent Research (Medical Sciences; grant 271-09-0187). C.L. was supported by a European Molecular Biology Organization postdoctoral fellowship. This research was further supported by grants from the Swedish Research council, the Swedish Cancer Society and the integrated EU project European drug initiative on channels and transporters (EDICT), as well as a Singapore National Research Foundation Competitive Research Programme grant (NRF2008NRF-CRP002-067). We thank Diamond Light Source for access to beamline I02 (MX5873 and MX6603) that contributed to the results presented here and acknowledge the SOLEIL synchrotron for provision of synchrotron radiation facilities at beamline PROXIMA1 (proposal 20110314). We also thank the Protein Science Facility at the Karolinska Institute for provision of crystallization infrastructure.
Author information
Authors and Affiliations
Contributions
E.M.Q., C.L. and P.M. conducted experiments. L.T. assisted in data collection and analysis. E.M.Q. and C.L. wrote the initial manuscript, and L.T. and P.N. contributed with revisions. P.N. supervised the project. All authors contributed to experimental design and have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Table 1 (PDF 2120 kb)
Rights and permissions
About this article
Cite this article
Quistgaard, E., Löw, C., Moberg, P. et al. Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters. Nat Struct Mol Biol 20, 766–768 (2013). https://doi.org/10.1038/nsmb.2569
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2569
This article is cited by
-
Molecular basis for inhibiting human glucose transporters by exofacial inhibitors
Nature Communications (2022)
-
GLUT3 inhibitor discovery through in silico ligand screening and in vivo validation in eukaryotic expression systems
Scientific Reports (2022)
-
The molecular basis for sugar import in malaria parasites
Nature (2020)
-
Protein structure reveals how a malaria parasite imports a wide range of sugars
Nature (2020)
-
Hydrogen-deuterium exchange mass spectrometry captures distinct dynamics upon substrate and inhibitor binding to a transporter
Nature Communications (2020)