Abstract
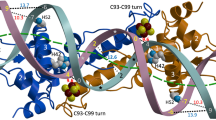
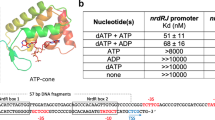
IscR from Escherichia coli is an unusual metalloregulator in that both apo and iron sulfur (Fe-S)-IscR regulate transcription and exhibit different DNA binding specificities. Here, we report structural and biochemical studies of IscR suggesting that remodeling of the protein-DNA interface upon Fe-S ligation broadens the DNA binding specificity of IscR from binding the type 2 motif only to both type 1 and type 2 motifs. Analysis of an apo-IscR variant with relaxed target-site discrimination identified a key residue in wild-type apo-IscR that, we propose, makes unfavorable interactions with a type 1 motif. Upon Fe-S binding, these interactions are apparently removed, thereby allowing holo-IscR to bind both type 1 and type 2 motifs. These data suggest a unique mechanism of ligand-mediated DNA site recognition, whereby metallocluster ligation relocates a protein-specificity determinant to expand DNA target-site selection, allowing a broader transcriptomic response by holo-IscR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Johnson, D.C., Dean, D.R., Smith, A.D. & Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 (2005).
Fleischhacker, A.S. & Kiley, P.J. Iron-containing transcription factors and their roles as sensors. Curr. Opin. Chem. Biol. 15, 335–341 (2011).
Crack, J.C., Green, J., Thomson, A.J. & Le Brun, N.E. Iron-sulfur cluster sensor-regulators. Curr. Opin. Chem. Biol. 16, 35–44 (2012).
Guerra, A.J. & Giedroc, D.P. Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch. Biochem. Biophys. 519, 210–222 (2012).
Fleischhacker, A.S. et al. Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR. Biochemistry 51, 4453–4462 (2012).
Nesbit, A.D., Giel, J.L., Rose, J.C. & Kiley, P.J. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J. Mol. Biol. 387, 28–41 (2009).
Yeo, W.S., Lee, J.H., Lee, K.C. & Roe, J.H. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 61, 206–218 (2006).
Giel, J.L. et al. Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli. Mol. Microbiol. 87, 478–492 (2013).
Rodionov, D.A., Gelfand, M.S., Todd, J.D., Curson, A.R. & Johnston, A.W. Computational reconstruction of iron- and manganese-responsive transcriptional networks in α-proteobacteria. PLoS Comput. Biol. 2, e163 (2006).
Shepard, W. et al. Insights into the Rrf2 repressor family–the structure of CymR, the global cysteine regulator of Bacillus subtilis. FEBS J. 278, 2689–2701 (2011).
Schwartz, C.J. et al. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98, 14895–14900 (2001).
Giel, J.L., Rodionov, D., Liu, M., Blattner, F.R. & Kiley, P.J. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 60, 1058–1075 (2006).
Lazazzera, B.A., Beinert, H., Khoroshilova, N., Kennedy, M.C. & Kiley, P.J. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271, 2762–2768 (1996).
Hidalgo, E. & Demple, B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 13, 138–146 (1994).
Gaudu, P. & Weiss, B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc. Natl. Acad. Sci. USA 93, 10094–10098 (1996).
Ebright, R.H. et al. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc. Natl. Acad. Sci. USA 84, 6083–6087 (1987).
Komori, H. et al. Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6-Å resolution. EMBO J. 18, 4597–4607 (1999).
MacPherson, S., Larochelle, M. & Turcotte, B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70, 583–604 (2006).
Reeves, R. & Nissen, M.S. The A•T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 265, 8573–8582 (1990).
Rohs, R. et al. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 79, 233–269 (2010).
Rohs, R. et al. The role of DNA shape in protein-DNA recognition. Nature 461, 1248–1253 (2009).
Haran, T.E. & Mohanty, U. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 42, 41–81 (2009).
Coulocheri, S.A., Pigis, D.G., Papavassiliou, K.A. & Papavassiliou, A.G. Hydrogen bonds in protein-DNA complexes: where geometry meets plasticity. Biochimie 89, 1291–1303 (2007).
Ding, H., Hidalgo, E. & Demple, B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J. Biol. Chem. 271, 33173–33175 (1996).
Watanabe, S., Kita, A., Kobayashi, K. & Miki, K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc. Natl. Acad. Sci. USA 105, 4121–4126 (2008).
Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59, 1073–1082 (2006).
Nesbit, A.D., Fleischhacker, A.S., Teter, S.J. & Kiley, P.J. ArcA and AppY antagonize IscR repression of hydrogenase 1 expression under anaerobic conditions, revealing a novel mode of O2 regulation of gene expression in Escherichia coli. J. Bacteriol. 194, 6892–6899 (2012).
Shin, J.-H. et al. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc. Natl. Acad. Sci. USA 108, 5045–5050 (2011).
Ohmura, T., Ueda, T., Hashimoto, Y. & Imoto, T. Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme. Protein Eng. 14, 421–425 (2001).
Leahy, D.J., Erickson, H.P., Aukhil, I., Joshi, P. & Hendrickson, W.A. Crystallization of a fragment of human fibronectin: introduction of methionine by site-directed mutagenesis to allow phasing via selenomethionine. Proteins 19, 48–54 (1994).
Battye, T.G., Kontogiannis, L., Johnson, O., Powell, H.R. & Leslie, A.G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Painter, J. & Merritt, E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 (2006).
McNicholas, S., Potterton, E., Wilson, K.S. & Noble, M.E. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 (2011).
Fischer, D.S. & Price, D.C. A simple serum iron method using the new sensitive chromogen tripyridyl-s-triazine. Clin. Chem. 10, 21–31 (1964).
Acknowledgements
We thank the organizers and instructors of the 2012 CCP4 APS school (attended by S.R.) for providing valuable insights into analyzing the data. Structural data were collected at the Berkeley Center for Structural Biology (BCSB) beamlines at Advanced Light Source (ALS) and at the Northeastern Collaborative Access Team (NE-CAT) beamlines at Advanced Photon Source (APS). BCSB is supported in part by the NIH, the National Institute of General Medical Sciences and the Howard Hughes Medical Institute. The ALS is supported by the Director, Office of Science and the Office of Basic Energy Sciences of the US Department of Energy (DOE) under contract no. DE-AC02-05CH11231. The NE-CAT beamlines are supported by grants from the National Center for Research Resources (5P41RR015301-10) and the National Institute of General Medical Sciences (8 P41 GM103403-10). Use of the APS, operated for the DOE Office of Science by Argonne National Laboratory, was supported by the DOE under contract no. DE-AC02-06CH11357. This work was funded by grants from the US National Institutes of Health (NIH; GM045844 to P.J.K.) and The Methodist Hospital Research Institute to K.J.P.
Author information
Authors and Affiliations
Contributions
P.J.K., K.J.P., S.J.T., S.R. and R.G.B. designed the experiments. S.R., S.J.T. and P.H.Z. performed the experiments. P.J.K., K.J.P., S.J.T. and S.R. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Table 1 (PDF 15284 kb)
Rights and permissions
About this article
Cite this article
Rajagopalan, S., Teter, S., Zwart, P. et al. Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity. Nat Struct Mol Biol 20, 740–747 (2013). https://doi.org/10.1038/nsmb.2568
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2568
This article is cited by
-
Structural determinants of DNA recognition by the NO sensor NsrR and related Rrf2-type [FeS]-transcription factors
Communications Biology (2022)
-
Structure and functional implications of WYL domain-containing bacterial DNA damage response regulator PafBC
Nature Communications (2019)
-
Crystal structures of the NO sensor NsrR reveal how its iron-sulfur cluster modulates DNA binding
Nature Communications (2017)
-
Metal homeostasis and resistance in bacteria
Nature Reviews Microbiology (2017)
-
Selenite reduction by the obligate aerobic bacterium Comamonas testosteroni S44 isolated from a metal-contaminated soil
BMC Microbiology (2014)