Abstract
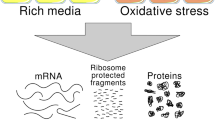
Proteins regulate gene expression by controlling mRNA biogenesis, localization, translation and decay. Identifying the composition, diversity and function of mRNA–protein complexes (mRNPs) is essential to understanding these processes. In a global survey of Saccharomyces cerevisiae mRNA-binding proteins, we identified 120 proteins that cross-link to mRNA, including 66 new mRNA-binding proteins. These include kinases, RNA-modification enzymes, metabolic enzymes and tRNA- and rRNA-metabolism factors. These proteins show dynamic subcellular localization during stress, including assembly into stress granules and processing bodies (P bodies). Cross-linking and immunoprecipitation (CLIP) analyses of the P-body components Pat1, Lsm1, Dhh1 and Sbp1 identified sites of interaction on specific mRNAs, revealing positional binding preferences and co-assembly preferences. When taken together, this work defines the major yeast mRNP proteins, reveals widespread changes in their subcellular location during stress and begins to define assembly rules for P-body mRNPs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Tsvetanova, N.G., Klass, D.M., Salzman, J. & Brown, P.O. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS ONE 5, e12671 (2010).
Matunis, M.J., Matunis, E.L. & Dreyfuss, G. PUB1: a major yeast poly(A)+ RNA-binding protein. Mol. Cell. Biol. 13, 6114–6123 (1993).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Baltz, A.G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M. & Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382 (2005).
Holcik, M. & Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 (2005).
Dewey, C.M. et al. TDP-43 aggregation in neurodegeneration: Are stress granules the key? Brain Res. 10.1016/j.brainres.2012.02.032 (2012).
Nogueira, T., de Smit, M., Graffe, M. & Springer, M. The relationship between translational control and mRNA degradation for the Escherichia coli threonyl-tRNA synthetase gene. J. Mol. Biol. 310, 709–722 (2001).
Frugier, M. & Giegé, R. Yeast aspartyl-tRNA synthetase binds specifically its own mRNA. J. Mol. Biol. 331, 375–383 (2003).
Sampath, P. et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119, 195–208 (2004).
Hogan, D.J., Riordan, D.P., Gerber, A.P., Herschlag, D. & Brown, P.O. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6, e255 (2008).
Du, T.-G. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 9, 781–787 (2008).
Decker, C.J., Teixeira, D. & Parker, R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437–449 (2007).
Buchan, J.R. & Parker, R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009).
Buchan, J.R., Yoon, J.-H. & Parker, R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124, 228–239 (2011).
Hoyle, N.P., Castelli, L.M., Campbell, S.G., Holmes, L.E.A. & Ashe, M.P. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179, 65–74 (2007).
Grousl, T. et al. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122, 2078–2088 (2009).
Sheth, U. & Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808 (2003).
Nissan, T., Rajyaguru, P., She, M., Song, H. & Parker, R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 39, 773–783 (2010).
Segal, S.P., Dunckley, T. & Parker, R. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 5120–5130 (2006).
Chowdhury, A., Mukhopadhyay, J. & Tharun, S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13, 998–1016 (2007).
He, W. & Parker, R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158, 1445–1455 (2001).
Bailey, T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659 (2011).
Rajyaguru, P., She, M. & Parker, R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 45, 244–254 (2012).
Hoell, J.I. et al. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 10.1038/nsmb.2163 (2011).
Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A. & Parker, R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7, 1717–1727 (2001).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Kraft, C., Deplazes, A., Sohrmann, M. & Peter, M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610 (2008).
Yoon, J.-H., Choi, E.-J. & Parker, R. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189, 813–827 (2010).
Tharun, S. & Parker, R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8, 1075–1083 (2001).
Hilgers, V., Teixeira, D. & Parker, R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA 12, 1835–1845 (2006).
Coller, J. & Parker, R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73, 861–890 (2004).
Swisher, K.D. & Parker, R. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS ONE 5, e10006 (2010).
Teixeira, D. & Parker, R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2274–2287 (2007).
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996).
Andon, N.L. et al. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics 2, 1156–1168 (2002).
Qian, W.-J. et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res. 4, 53–62 (2005).
Buchan, J.R., Muhlrad, D. & Parker, R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441–455 (2008).
Ule, J., Jensen, K., Mele, A. & Darnell, R.B. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37, 376–386 (2005).
Wang, Z., Tollervey, J., Briese, M., Turner, D. & Ule, J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods 48, 287–293 (2009).
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008).
Bailey, T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659 (2011).
Acknowledgements
MS and proteomics data were acquired by the Arizona Proteomics Consortium supported by the US National Institute of Environmental Health Science grant ES06694 (the S.W.E.H.S.C.), the US National Institutes of Health, US National Cancer Institute grant CA023074 (the A.Z.C.C.) and the BIO5 Institute of the University of Arizona. The Thermo Fisher LTQ Orbitrap Velos mass spectrometer was provided by the US National Institutes of Health, US National Center for Research Resources grant 1S10 RR028868-01 (G.T.). We thank J.R. Buchan for assistance with microscopy and C. Decker and other members of the Parker lab for helpful discussions. This work was supported by funding from the US National Institutes of Health grant 7R37 GM045443 (R.P.), a Howard Hughes Medical Institute grant (R.P.) and Leukemia and Lymphoma Society fellowship 5687-13 (S.F.M.).
Author information
Authors and Affiliations
Contributions
R.P., S.F.M., S.J. and M.S. designed the project. S.F.M. performed the in vivo RBP capture experiments and CLIP analysis, S.J. did the microscopy and CLIP analysis, and M.S. performed the CLIP experiments and analysis. R.P., S.F.M. and S.J. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Table 2, Supplementary note (PDF 378 kb)
Supplementary Table 1
A list of all 120 proteins identified as enriched by the in vivo capture of RBPs. (XLSX 48 kb)
Supplementary Table 3
A list of mRNAs (Tier1 and Tier2) bound by Dhh1, Lsm1, Pat1 and Sbp1. (XLSX 127 kb)
Rights and permissions
About this article
Cite this article
Mitchell, S., Jain, S., She, M. et al. Global analysis of yeast mRNPs. Nat Struct Mol Biol 20, 127–133 (2013). https://doi.org/10.1038/nsmb.2468
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2468
This article is cited by
-
Lsm7 phase-separated condensates trigger stress granule formation
Nature Communications (2022)
-
Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains
Nature Chemistry (2022)
-
Clueless/CLUH regulates mitochondrial fission by promoting recruitment of Drp1 to mitochondria
Nature Communications (2022)
-
A translation enhancer element from black beetle virus engages yeast eIF4G1 to drive cap-independent translation initiation
Scientific Reports (2021)
-
The rate and molecular spectrum of mutation are selectively maintained in yeast
Nature Communications (2021)