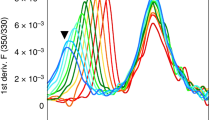
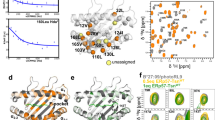
Abstract
Mammalian class II major histocompatibility (MHCII) proteins bind peptide antigens in endosomal compartments of antigen-presenting cells. The nonclassical MHCII protein HLA-DM chaperones peptide-free MHCII, protecting it against inactivation, and catalyzes peptide exchange on loaded MHCII. Another nonclassical MHCII protein, HLA-DO, binds HLA-DM and influences the repertoire of peptides presented by MHCII proteins. However, the mechanism by which HLA-DO functions is unclear. Here we have used X-ray crystallography, enzyme kinetics and mutagenesis approaches to investigate human HLA-DO structure and function. In complex with HLA-DM, HLA-DO adopts a classical MHCII structure, with alterations near the α subunit's 310 helix. HLA-DO binds to HLA-DM at the same sites implicated in MHCII interaction, and kinetic analysis showed that HLA-DO acts as a competitive inhibitor. These results show that HLA-DO inhibits HLA-DM function by acting as a substrate mimic, and the findings also limit the possible functional roles for HLA-DO in antigen presentation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Denzin, L.K. & Cresswell, P. HLA-DM induces CLIP dissociation from MHC class II αβ dimers and facilitates peptide loading. Cell 82, 155–165 (1995).
Sloan, V.S. et al. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature 375, 802–806 (1995).
Ghosh, P., Amaya, M., Mellins, E. & Wiley, D.C. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature 378, 457–462 (1995).
Morris, P. et al. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature 368, 551–554 (1994).
Riberdy, J.M., Newcomb, J.R., Surman, M.J., Barbosa, J.A. & Cresswell, P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature 360, 474–477 (1992).
Anders, A.K. et al. HLA-DM captures partially empty HLA-DR molecules for catalyzed removal of peptide. Nat. Immunol. 12, 54–61 (2011).
Ferrante, A. & Gorski, J. Cutting edge: HLA-DM-mediated peptide exchange functions normally on MHC class II-peptide complexes that have been weakened by elimination of a conserved hydrogen bond. J. Immunol. 184, 1153–1158 (2010).
Narayan, K. et al. HLA-DM targets the hydrogen bond between the histidine at position β81 and peptide to dissociate HLA-DR-peptide complexes. Nat. Immunol. 8, 92–100 (2007).
Painter, C. et al. Conformational lability in the class II MHC 3-10 helix and adjacent extended strand dictate HLA-DM susceptibility and peptide exchange. Proc. Natl. Acad. Sci. USA 108, 19329–19334 (2011).
Zhou, Z., Callaway, K.A., Weber, D.A. & Jensen, P.E. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J. Immunol. 183, 4187–4191 (2009).
Denzin, L.K., Hammond, C. & Cresswell, P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J. Exp. Med. 184, 2153–2165 (1996).
Kropshofer, H., Arndt, S.O., Moldenhauer, G., Hammerling, G.J. & Vogt, A.B. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity 6, 293–302 (1997).
Weber, D.A., Evavold, B.D. & Jensen, P.E. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science 274, 618–620 (1996).
Tonnelle, C., DeMars, R. & Long, E.O. DOβ: a new β chain gene in HLA-D with a distinct regulation of expression. EMBO J. 4, 2839–2847 (1985).
Douek, D.C. & Altmann, D.M. T-cell apoptosis and differential human leucocyte antigen class II expression in human thymus. Immunology 99, 249–256 (2000).
Hornell, T.M. et al. Human dendritic cell expression of HLA-DO is subset specific and regulated by maturation. J. Immunol. 176, 3536–3547 (2006).
Karlsson, L., Surh, C.D., Sprent, J. & Peterson, P.A. A novel class II MHC molecule with unusual tissue distribution. Nature 351, 485–488 (1991).
Chen, X. et al. Regulated expression of human histocompatibility leukocyte antigen (HLA)-DO during antigen-dependent and antigen-independent phases of B cell development. J. Exp. Med. 195, 1053–1062 (2002).
Glazier, K.S. et al. Germinal center B cells regulate their capability to present antigen by modulation of HLA-DO. J. Exp. Med. 195, 1063–1069 (2002).
Chen, X., Reed-Loisel, L.M., Karlsson, L. & Jensen, P.E. H2-O expression in primary dendritic cells. J. Immunol. 176, 3548–3556 (2006).
Denzin, L.K., Fallas, J.L., Prendes, M. & Yi, W. Right place, right time, right peptide: DO keeps DM focused. Immunol. Rev. 207, 279–292 (2005).
Jensen, P.E. Antigen processing: HLA-DO—a hitchhiking inhibitor of HLA-DM. Curr. Biol. 8, R128–R131 (1998).
Yi, W. et al. Targeted regulation of self-peptide presentation prevents type I diabetes in mice without disrupting general immunocompetence. J. Clin. Invest. 120, 1324–1336 (2010).
Perraudeau, M. et al. Altered major histocompatibility complex class II peptide loading in H2-O-deficient mice. Eur. J. Immunol. 30, 2871–2880 (2000).
van Ham, S.M. et al. HLA-DO is a negative modulator of HLA-DM-mediated MHC class II peptide loading. Curr. Biol. 7, 950–957 (1997).
Alfonso, C. et al. Analysis of H2-O influence on antigen presentation by B cells. J. Immunol. 171, 2331–2337 (2003).
Alfonso, C., Williams, G.S. & Karlsson, L. H2-O influence on antigen presentation in H2-E-expressing mice. Eur. J. Immunol. 33, 2014–2021 (2003).
Liljedahl, M. et al. Altered antigen presentation in mice lacking H2-O. Immunity 8, 233–243 (1998).
Denzin, L.K., Sant'Angelo, D.B., Hammond, C., Surman, M.J. & Cresswell, P. Negative regulation by HLA-DO of MHC class II-restricted antigen processing. Science 278, 106–109 (1997).
Liljedahl, M. et al. HLA-DO is a lysosomal resident which requires association with HLA-DM for efficient intracellular transport. EMBO J. 15, 4817–4824 (1996).
Yoon, T. et al. Mapping the DO/DM complex by FRET and mutagenesis. Proc. Natl. Acad. Sci. USA 109, 11276–11281 (2012).
Fallas, J.L. et al. Ectopic expression of HLA-DO in mouse dendritic cells diminishes MHC class II antigen presentation. J. Immunol. 173, 1549–1560 (2004).
Thibodeau, J. et al. Conserved structural features between HLA-DOβ and -DRβ. Mol. Immunol. 35, 885–893 (1998).
Kropshofer, H. et al. A role for HLA-DO as a co-chaperone of HLA-DM in peptide loading of MHC class II molecules. EMBO J. 17, 2971–2981 (1998).
Deshaies, F. et al. A point mutation in the groove of HLA-DO allows egress from the endoplasmic reticulum independent of HLA-DM. Proc. Natl. Acad. Sci. USA 102, 6443–6448 (2005).
Fremont, D.H., Crawford, F., Marrack, P., Hendrickson, W.A. & Kappler, J. Crystal structure of mouse H2-M. Immunity 9, 385–393 (1998).
Mosyak, L., Zaller, D.M. & Wiley, D.C. The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation. Immunity 9, 377–383 (1998).
Nicholson, M.J. et al. Small molecules that enhance the catalytic efficiency of HLA-DM. J. Immunol. 176, 4208–4220 (2006).
Pashine, A. et al. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity 19, 183–192 (2003).
Doebele, R.C., Busch, R., Scott, H.M., Pashine, A. & Mellins, E.D. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity 13, 517–527 (2000).
Vogt, A.B., Kropshofer, H., Moldenhauer, G. & Hammerling, G.J. Kinetic analysis of peptide loading onto HLA-DR molecules mediated by HLA-DM. Proc. Natl. Acad. Sci. USA 93, 9724–9729 (1996).
Copeland, R. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis 2nd edn., Ch. 9 (Wiley-VCH, 2000).
Williams, J.W. & Morrison, J.F. The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63, 437–467 (1979).
Zwart, W. et al. Spatial separation of HLA-DM/HLA-DR interactions within MIIC and phagosome-induced immune escape. Immunity 22, 221–233 (2005).
van Ham, M. et al. Modulation of the major histocompatibility complex class II-associated peptide repertoire by human histocompatibility leukocyte antigen (HLA)-DO. J. Exp. Med. 191, 1127–1136 (2000).
van Lith, M. et al. Regulation of MHC class II antigen presentation by sorting of recycling HLA-DM/DO and class II within the multivesicular body. J. Immunol. 167, 884–892 (2001).
Xiu, F. et al. Cutting edge: HLA-DO impairs the incorporation of HLA-DM into exosomes. J. Immunol. 187, 1547–1551 (2011).
Gondré-Lewis, T.A., Moquin, A.E. & Drake, J.R. Prolonged antigen persistence within nonterminal late endocytic compartments of antigen-specific B lymphocytes. J. Immunol. 166, 6657–6664 (2001).
Sant, A.J., Braunstein, N.S. & Germain, R.N. Predominant role of amino-terminal sequences in dictating efficiency of class II major histocompatibility complex αβ dimer expression. Proc. Natl. Acad. Sci. USA 84, 8065–8069 (1987).
Sant, A.J., Hendrix, L.R., Coligan, J.E., Maloy, W.L. & Germain, R.N. Defective intracellular transport as a common mechanism limiting expression of inappropriately paired class II major histocompatibility complex α/β chains. J. Exp. Med. 174, 799–808 (1991).
Rinderknecht, C.H. et al. DM influences the abundance of major histocompatibility complex class II alleles with low affinity for class II-associated invariant chain peptides via multiple mechanisms. Immunology 131, 18–32 (2010).
Ferrante, A., Anderson, M.W., Klug, C.S. & Gorski, J. HLA-DM mediates epitope selection by a “compare-exchange” mechanism when a potential peptide pool is available. PLoS ONE 3, e3722 (2008).
Painter, C.A., Cruz, A., Lopez, G.E., Stern, L.J. & Zavala-Ruiz, Z. Model for the peptide-free conformation of class II MHC proteins. PLoS ONE 3, e2403 (2008).
Weiss, M.S. Global indicators of X-ray data quality. J. Appl. Crystallogr. 34, 130–135 (2001).
Busch, R., Doebele, R.C., von Scheven, E., Fahrni, J. & Mellins, E.D. Aberrant intermolecular disulfide bonding in a mutant HLA-DM molecule: implications for assembly, maturation, and function. J. Immunol. 160, 734–743 (1998).
Marsh, S.G.E. et al. Nomenclature for factors of the HLA system. Tissue Antigens 75, 291–455 (2010).
Frayser, M., Sato, A.K., Xu, L. & Stern, L.J. Empty and peptide-loaded class II major histocompatibility complex proteins produced by expression in Escherichia coli and folding in vitro. Protein Expr. Purif. 15, 105–114 (1999).
Joshi, R.V., Zarutskie, J.A. & Stern, L.J. A three-step kinetic mechanism for peptide binding to MHC class II proteins. Biochemistry 39, 3751–3762 (2000).
Zarutskie, J.A. et al. The kinetic basis of peptide exchange catalysis by HLA-DM. Proc. Natl. Acad. Sci. USA 98, 12450–12455 (2001).
Otwinoski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Sheldrick, G.M. & Schneider, T.R. SHELXL: high-resolution refinement. Methods Enzymol. 277, 319–343 (1997).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Karplus, P.A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012).
Acknowledgements
We thank L. Karlsson (Johnson & Johnson) for S2 cells expressing DM and DO-Fc, J. Thibodeau (Université de Montréal) for plasmid pRmHa3 encoding sDOα and sDOβ cDNAs, L. Lee and L. Lu for assistance with insect cell culture and protein purification, H. Robinson and A. Heroux for assistance with crystallographic data collection, and E. Stratikos for helpful comments. Data for this study were measured at beamlines X25 and X29 of the US National Synchrotron Light Source, with support from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy and from the National Center for Research Resources (NCRR) of the US National Institutes of Health (NIH). This work was supported by NIH grants AI-38996 (L.J.S.), AI-48833 (L.J.S.), T32 AI07349 (S.E.M.), F32 AI072984 (S.E.M.), AI-095813 (E.D.M.) and AI-075253 (E.D.M.), by NIH/NCRR Clinical and Translational Science Award UL1 RR025744 at Stanford University, and by the Lucile Packard Foundation for Children's Health (E.D.M.).
Author information
Authors and Affiliations
Contributions
S.E.M. prepared and crystallized the DO–DM complex; S.E.M., A.I.G. and L.J.S. interpreted diffraction data; A.I.G. built the structural model; S.E.M. performed kinetics experiments; T.Y., W.J. and A.I.G. analyzed mutant proteins; A.I.G., S.E.M., T.Y., C.A.P., W.J., E.D.M. and L.J.S. designed experiments and analyzed data; A.I.G., S.E.M., C.A.P., W.J., E.D.M. and L.J.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Table 1 and Supplementary Figures 1–4 (PDF 3058 kb)
Rights and permissions
About this article
Cite this article
Guce, A., Mortimer, S., Yoon, T. et al. HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nat Struct Mol Biol 20, 90–98 (2013). https://doi.org/10.1038/nsmb.2460
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2460
This article is cited by
-
MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange
Nature Chemical Biology (2023)
-
Distinct editing functions of natural HLA-DM allotypes impact antigen presentation and CD4+ T cell activation
Cellular & Molecular Immunology (2020)
-
Dynamic plasticity of the lipid antigen-binding site of CD1d is crucially favoured by acidic pH and helper proteins
Scientific Reports (2020)
-
Synergy between B cell receptor/antigen uptake and MHCII peptide editing relies on HLA-DO tuning
Scientific Reports (2019)
-
What to do with HLA-DO/H-2O two decades later?
Immunogenetics (2019)