Abstract
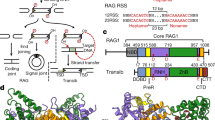
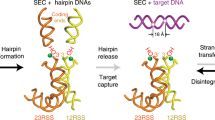
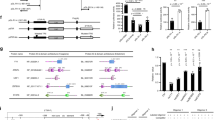
The formation of diverse immunoglobulin genes results in part from Rag protein–mediated DNA double-strand breaks at the edges of immunoglobulin gene segments, followed by combinatorial reassembly of these segments. We report that a Transib transposase from the insect Helicoverpa zea is active in vitro and that its breakage and joining activities mimic those of Rag, providing strong evidence that Rag and Transib transposases were derived from a common progenitor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Matthews, A.G. & Oettinger, M.A. Nat. Immunol. 10, 817–821 (2009).
Schatz, D.G. & Swanson, P.C. Annu. Rev. Genet. 45, 167–202 (2011).
Hiom, K., Melek, M. & Gellert, M. Cell 94, 463–470 (1998).
Agrawal, A., Eastman, Q.M. & Schatz, D.G. Nature 394, 744–751 (1998).
Kapitonov, V.V. & Jurka, J. PLoS Biol. 3, e181 (2005).
Chen, S. & Li, X. Gene 408, 51–63 (2008).
Du, E., Ni, X., Zhao, H. & Li, X. Insect Mol. Biol. 20, 291–301 (2011).
Hickman, A.B., Chandler, M. & Dyda, F. Crit. Rev. Biochem. Mol. Biol. 45, 50–69 (2010).
Kim, D.R. et al. Genes Dev. 13, 3070–3080 (1999).
Landree, M.A., Wibbenmeyer, J.A. & Roth, D.B. Genes Dev. 13, 3059–3069 (1999).
Fugmann, S.D., Villey, I.J., Ptaszek, L.M. & Schatz, D.G. Mol. Cell 5, 97–107 (2000).
Zhou, L. et al. Nature 432, 995–1001 (2004).
Yang, W., Lee, J.Y. & Nowotny, M. Mol. Cell 22, 5–13 (2006).
Acknowledgements
We thank the other members of the Craig lab for fruitful discussions, H. McComas for her assistance with the figures and text, and S. Desiderio for his comments on the manuscript. N.L.C. is supported as a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
N.L.C. conceived the project; C.G.H., X.L. and N.L.C. designed the experiments; C.G.H. and X.L. carried out the experiments; C.G.H., X.L. and N.L.C. analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 1507 kb)
Rights and permissions
About this article
Cite this article
Hencken, C., Li, X. & Craig, N. Functional characterization of an active Rag-like transposase. Nat Struct Mol Biol 19, 834–836 (2012). https://doi.org/10.1038/nsmb.2338
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2338
This article is cited by
-
Structural insights into the evolution of the RAG recombinase
Nature Reviews Immunology (2022)
-
Identification of RAG-like transposons in protostomes suggests their ancient bilaterian origin
Mobile DNA (2020)
-
Structural basis of seamless excision and specific targeting by piggyBac transposase
Nature Communications (2020)
-
Transposable elements in human genetic disease
Nature Reviews Genetics (2019)
-
Structures of a RAG-like transposase during cut-and-paste transposition
Nature (2019)