Abstract
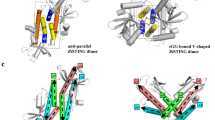
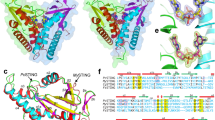
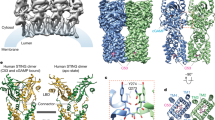
STING (stimulator of interferon genes) is an innate immune sensor of cyclic dinucleotides that regulates the induction of type I interferons. STING's C-terminal domain forms a V-shaped dimer and binds a cyclic diguanylate monophosphate (c-di-GMP) at the dimer interface by both direct and solvent-mediated hydrogen bonds. Guanines of c-di-GMP stack against the phenolic rings of a conserved tyrosine, and mutations at the c-di-GMP binding surface reduce nucleotide binding and affect signaling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Keating, S.E., Baran, M. & Bowie, A.G. Trends Immunol. 32, 574–581 (2011).
Kato, H., Takahasi, K. & Fujita, T. Immunol. Rev. 243, 91–98 (2011).
McWhirter, S.M. et al. J. Exp. Med. 206, 1899–1911 (2009).
Woodward, J.J., Iavarone, A.T. & Portnoy, D.A. Science 328, 1703–1705 (2010).
Karaolis, D.K. et al. J. Immunol. 178, 2171–2181 (2007).
Sondermann, H., Shikuma, N.J. & Yildiz, F.H. Curr. Opin. Microbiol. 15, 140–146 (2012).
Pesavento, C. & Hengge, R. Curr. Opin. Microbiol. 12, 170–176 (2009).
Burdette, D.L. et al. Nature 478, 515–518 (2011).
Barber, G.N. Curr. Opin. Immunol. 23, 10–20 (2011).
Barber, G.N. Nat. Immunol. 12, 929–930 (2011).
Unterholzner, L. et al. Nat. Immunol. 11, 997–1004 (2010).
Zhang, Z. et al. Nat. Immunol. 12, 959–965 (2011).
Tanaka, Y. & Chen, Z.J. Sci. Signal. 5, ra20 (2012).
Ye, H., Park, Y.C., Kreishman, M., Kieff, E. & Wu, H. Mol. Cell 4, 321–330 (1999).
Pai, E.F. et al. Nature 341, 209–214 (1989).
Ishikawa, H. & Barber, G.N. Nature 455, 674–678 (2008).
Ouyang, S. et al. Immunity published online, doi:10.1016/j.immuni.2012.03.019 (10 May 2012).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1997).
Sheldrick, G.M. Acta Crystallogr. A 64, 112–122 (2008).
Adams, P.D. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Kabsch, W. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010).
Winn, M.D. et al. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
tenOever, B.R. et al. J. Virol. 78, 10636–10649 (2004).
Acknowledgements
The diffraction data of the SeMet crystals were collected at beamline 11.1 at the Stanford Synchrotron Radiation Lightsource (SSRL). The ITC binding studies were conducted in T. Begley's laboratory. This research is supported by the US National Institute of Health (grants AI087741 to P.L. and AI073335 to C.C.K.).
Author information
Authors and Affiliations
Contributions
C.S. crystallized the protein and conducted the binding and kinase assays. P.L. determined the structures. G.Y. conducted the IFN-β reporter assays. T.W. helped with the data collection. P.L., C.C.K., C.S. and T.W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Table 1 (PDF 4674 kb)
Rights and permissions
About this article
Cite this article
Shu, C., Yi, G., Watts, T. et al. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol 19, 722–724 (2012). https://doi.org/10.1038/nsmb.2331
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2331
This article is cited by
-
cGAMP-activated cGAS–STING signaling: its bacterial origins and evolutionary adaptation by metazoans
Nature Structural & Molecular Biology (2023)
-
Crystal structure and functional implication of bacterial STING
Nature Communications (2022)
-
ICA69 aggravates ferroptosis causing septic cardiac dysfunction via STING trafficking
Cell Death Discovery (2022)
-
The roles of transmembrane family proteins in the regulation of store-operated Ca2+ entry
Cellular and Molecular Life Sciences (2022)
-
STING cyclic dinucleotide sensing originated in bacteria
Nature (2020)