Abstract
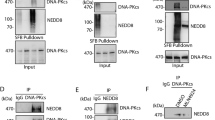
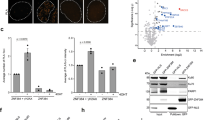
The ubiquitination cascade has a key role in the assembly of repair and signaling proteins at sites of double-strand DNA breaks. The E3 ubiquitin ligase RING finger protein 8 (RNF8) triggers the initial ubiquitination at double-strand DNA breaks, whereas sustained ubiquitination requires the downstream E3 ligase RING finger protein 168 (RNF168). It is not known whether RNF8 and RNF168 have discrete substrates and/or form different ubiquitin chains. Here we show that RNF168 acts with the ubiquitin-conjugating enzyme E2 13 (UBC13) and specifically synthesizes Lys63-linked chains, whereas RNF8 primarily forms Lys48-linked chains on chromatin, which promote substrate degradation. We also find that RNF8 regulates the abundance of the nonhomologous end-joining (NHEJ) repair protein KU80 at sites of DNA damage, and that RNF8 depletion results in prolonged retention of KU80 at damage sites and impaired nonhomologous end-joining repair. These findings reveal a distinct feature of RNF8 and indicate the involvement of the ubiquitination-mediated degradation pathway in DNA damage repair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Weissman, A.M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2, 169–178 (2001).
Komander, D. et al. Molecular discrimination of structurally equivalent Lys63–linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 (2009).
Eddins, M.J., Varadan, R., Fushman, D., Pickart, C.M. & Wolberger, C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 367, 204–211 (2007).
Chau, V. et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 (1989).
Matsumoto, M.L. et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 39, 477–484 (2010).
Raiborg, C. & Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 (2009).
Kirkin, V., McEwan, D.G., Novak, I. & Dikic, I. A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 (2009).
Ulrich, H.D. & Walden, H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 11, 479–489 (2010).
Hofmann, R.M. & Pickart, C.M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999).
Wu-Baer, F., Lagrazon, K., Yuan, W. & Baer, R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278, 34743–34746 (2003).
Nishikawa, H. et al. Mass spectrometric and mutational analyses reveal Lys-6–linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 279, 3916–3924 (2004).
Zhao, G.Y. et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25, 663–675 (2007).
Huen, M.S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Kolas, N.K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007).
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Stewart, G.S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Feng, L., Wang, J. & Chen, J. The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. J. Biol. Chem. 285, 30982–30988 (2010).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
Newton, K. et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 (2008).
Fujimuro, M., Sawada, H. & Yokosawa, H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 349, 173–180 (1994).
Kim, H., Chen, J. & Yu, X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 (2007).
Wang, B. et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 (2007).
Plans, V. et al. The RING finger protein RNF8 recruits UBC13 for lysine 63–based self polyubiquitylation. J. Cell. Biochem. 97, 572–582 (2006).
Lorick, K.L. et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96, 11364–11369 (1999).
Fang, S., Jensen, J.P., Ludwig, R.L., Vousden, K.H. & Weissman, A.M. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945–8951 (2000).
Postow, L. et al. Ku80 removal from DNA through double strand break–induced ubiquitylation. J. Cell Biol. 182, 467–479 (2008).
Burma, S., Chen, B.P. & Chen, D.J. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst.) 5, 1042–1048 (2006).
Kim, J.S. et al. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 170, 341–347 (2005).
Mari, P.O. et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA 103, 18597–18602 (2006).
Galanty, Y. et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 (2009).
Li, L. et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J. Exp. Med. 207, 983–997 (2010).
Shao, G. et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23, 740–754 (2009).
Feng, L., Huang, J. & Chen, J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 23, 719–728 (2009).
Wang, B., Hurov, K., Hofmann, K. & Elledge, S.J. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 23, 729–739 (2009).
Penengo, L. et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124, 1183–1195 (2006).
Lukas, C., Falck, J., Bartkova, J., Bartek, J. & Lukas, J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5, 255–260 (2003).
Walker, J.R., Corpina, R.A. & Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 (2001).
Antoni, L., Sodha, N., Collins, I. & Garrett, M.D. CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat. Rev. Cancer 7, 925–936 (2007).
Lok, G.T. et al. Differential regulation of RNF8-mediated Lys48- and Lys63-based poly-ubiquitylation. Nucleic Acids Res. 40, 196–205 (2012).
Bekker-Jensen, S. et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 12, 80–6; (suppl. pp 1–12).
Acknowledgements
We thank our colleagues in the Chen laboratory for insightful discussions and technical assistance. We also thank X. Zhang and H.P. Adams for technical assistance with the laser microirradiation. This work was supported in part by grants from the US National Institutes of Health (CA089239 and CA092312 to J.C.). J.C. is a recipient of an Era of Hope Scholar award from the US Department of Defense (W81XWH-05-1-0470) and is a member of the MD Anderson Cancer Center (CA016672).
Author information
Authors and Affiliations
Contributions
L.F. designed and carried out the experiments. J.C. advised on the design of the experiments. L.F. and J.C. were responsible for the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Methods. (PDF 5149 kb)
Rights and permissions
About this article
Cite this article
Feng, L., Chen, J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 19, 201–206 (2012). https://doi.org/10.1038/nsmb.2211
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2211
This article is cited by
-
Profiling ubiquitin signalling with UBIMAX reveals DNA damage- and SCFβ-Trcp1-dependent ubiquitylation of the actin-organizing protein Dbn1
Nature Communications (2023)
-
A functional reference map of the RNF8 interactome in cancer
Biology Direct (2022)
-
Chidamide and venetoclax synergistically exert cytotoxicity on multiple myeloma by upregulating BIM expression
Clinical Epigenetics (2022)
-
USP44 regulates irradiation-induced DNA double-strand break repair and suppresses tumorigenesis in nasopharyngeal carcinoma
Nature Communications (2022)
-
RNF166 plays a dual role for Lys63-linked ubiquitination and sumoylation of its target proteins
Journal of Neural Transmission (2022)