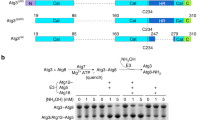
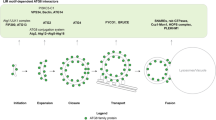
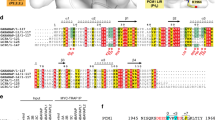
Abstract
Autophagy is the degradation of cellular organelles via the lysosomal pathway. The autophagic ubiquitin-like (Ubl) molecule Atg8 is activated by the E1-like enzyme Atg7. As this noncanonical E1 enzyme's domain organization is unique among Ubl-activating E1 enzymes, the structural basis for its interactions with Atg8 and partner E2 enzymes remains obscure. Here we present the structure of the N-terminal domain of Atg7, revealing a unique protein fold and interactions with both autophagic E2 enzymes Atg3 and Atg10. The structure of the C-terminal domain of Atg7 in complex with Atg8 shows the mode of dimerization and mechanism of recognition of Atg8. Notably, the catalytic cysteine residue in Atg7 is positioned close to the C-terminal glycine of Atg8, its target for thioester formation, potentially eliminating the need for large conformational rearrangements characteristic of other E1s.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nakatogawa, H., Suzuki, K., Kamada, Y. & Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 (2009).
Klionsky, D.J. & Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 (2000).
Klionsky, D.J. et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545 (2003).
Noda, N.N., Ohsumi, Y. & Inagaki, F. ATG systems from the protein structural point of view. Chem. Rev. 109, 1587–1598 (2009).
Suzuki, K. & Ohsumi, Y. Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett. 584, 1280–1286 (2010).
Shintani, T. & Klionsky, D.J. Autophagy in health and disease: a double-edged sword. Science 306, 990–995 (2004).
Rabinowitz, J.D. & White, E. Autophagy and metabolism. Science 330, 1344–1348 (2010).
Klionsky, D.J. et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy 6, 438–448 (2010).
Levine, B., Mizushima, N. & Virgin, H.W. Autophagy in immunity and inflammation. Nature 469, 323–335 (2011).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Varshavsky, A. Regulated protein degradation. Trends Biochem. Sci. 30, 283–286 (2005).
Hochstrasser, M. Origin and function of ubiquitin-like proteins. Nature 458, 422–429 (2009).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Schulman, B.A. & Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 (2009).
Lee, I. & Schindelin, H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 134, 268–278 (2008).
Huang, D.T. et al. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature 445, 394–398 (2007).
Olsen, S.K., Capili, A.D., Lu, X., Tan, D.S. & Lima, C.D. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912 (2010).
Völler, D. & Schindelin, H. And yet it moves: active site remodeling in the SUMO E1. Structure 18, 419–421 (2010).
Bacik, J.P., Walker, J.R., Ali, M., Schimmer, A.D. & Dhe-Paganon, S. Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: mechanistic insights into a minimalistic E1 enzyme. J. Biol. Chem. 285, 20273–20280 (2010).
Geng, J. & Klionsky, D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 9, 859–864 (2008).
Zhang, J. & Ney, P.A. Reticulocyte mitophagy: monitoring mitochondrial clearance in a mammalian model. Autophagy 6, 405–408 (2010).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Mortensen, M. et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 208, 455–467 (2011).
Lenz, H.D., Vierstra, R.D., Nurnberger, T. & Gust, A.A. ATG7 contributes to plant basal immunity towards fungal infection. Plant Signal. Behav. 6, 1040–1042 (2011).
Komatsu, M. et al. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J. Biol. Chem. 276, 9846–9854 (2001).
Noda, T., Fujita, N. & Yoshimori, T. The Ubi brothers reunited. Autophagy 4, 540–541 (2008).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Yamazaki-Sato, H., Tanida, I., Ueno, T. & Kominami, E. The carboxyl terminal 17 amino acids within Apg7 are essential for Apg8 lipidation, but not for Apg12 conjugation. FEBS Lett. 551, 71–77 (2003).
Lake, M.W., Wuebbens, M.M., Rajagopalan, K.V. & Schindelin, H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 414, 325–329 (2001).
Lehmann, C., Begley, T.P. & Ealick, S.E. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry 45, 11–19 (2006).
Lois, L.M. & Lima, C.D. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 (2005).
Harding, M.M. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr. D Biol. Crystallogr. 62, 678–682 (2006).
Reynolds, C., Damerell, D. & Jones, S. ProtorP: a protein-protein interaction analysis server. Bioinformatics 25, 413–414 (2009).
Hurley, J.H., Lee, S. & Prag, G. Ubiquitin-binding domains. Biochem. J. 399, 361–372 (2006).
Sato, Y. et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 (2008).
Yamada, Y. et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 282, 8036–8043 (2007).
Yamaguchi, M. et al. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 285, 29599–29607 (2010).
Noda, N.N., Ohsumi, Y. & Inagaki, F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385 (2010).
Ichimura, Y. et al. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283, 22847–22857 (2008).
Nakatogawa, H., Ichimura, Y. & Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 (2007).
Suzuki, K., Kubota, Y., Sekito, T. & Ohsumi, Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209–218 (2007).
Huang, W.P., Scott, S.V., Kim, J. & Klionsky, D.J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275, 5845–5851 (2000).
Hanada, T. et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 (2007).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Heinig, M. & Frishman, D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 32, W500–W502 (2004).
Acknowledgements
We thank the staff at 4A beamline, Pohang Accelerator Laboratory, South Korea, and NE3-A and NW12 beamline, Photon Factory, Japan, for help with the data collection; H. Nakatogawa and Y. Ohsumi (Tokyo Institute of Technology), W.-K. Huh (Seoul National University), C.-W. Yun (Korea University), D.J. Klionsky (University of Michigan) for yeast strains; and M.J. Eck for critical comments on the manuscript. This work was supported by the World-Class University Project (R33-10108), the 21C Frontier Functional Proteomics Project (FPR08B2-270) and the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A092006 and A084016).
Author information
Authors and Affiliations
Contributions
S.B.H., B.-W.K. and H.K.S. performed biochemical and structural studies; S.W.K. and J.K. performed yeast genetics; K.-E.L. and H.J. performed EM studies; S.B.H. and H.K.S. designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–8, Supplementary Tables 1 and 2 and Supplementary Methods (PDF 2589 kb)
Rights and permissions
About this article
Cite this article
Hong, S., Kim, BW., Lee, KE. et al. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat Struct Mol Biol 18, 1323–1330 (2011). https://doi.org/10.1038/nsmb.2165
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2165