Abstract
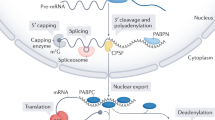
The Saccharomyces cerevisiae mRNA export adaptor Yra1 binds the Pcf11 subunit of cleavage-polyadenylation factor CF1A that links export to 3′ end formation. We found that an unexpected consequence of this interaction is that Yra1 influences cleavage-polyadenylation. Yra1 competes with the CF1A subunit Clp1 for binding to Pcf11, and excess Yra1 inhibits 3′ processing in vitro. Release of Yra1 at the 3′ ends of genes coincides with recruitment of Clp1, and depletion of Yra1 enhances Clp1 recruitment within some genes. These results suggest that CF1A is not necessarily recruited as a complete unit; instead, Clp1 can be incorporated co-transcriptionally in a process regulated by Yra1. Yra1 depletion causes widespread changes in poly(A) site choice, particularly at sites where the efficiency element is divergently positioned. We propose that one way Yra1 modulates cleavage-polyadenylation is by influencing co-transcriptional assembly of the CF1A 3′ processing factor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Iglesias, N. & Stutz, F. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 582, 1987–1996 (2008).
Schmid, M. & Jensen, T.H. Quality control of mRNP in the nucleus. Chromosoma 117, 419–429 (2008).
Luna, R., Gaillard, H., Gonzalez-Aguilera, C. & Aguilera, A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma 117, 319–331 (2008).
Perales, R. & Bentley, D. 'Cotranscriptionality': the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36, 178–191 (2009).
Eckner, R., Ellmeier, W. & Birnstiel, M.L. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 10, 3513–3522 (1991).
Huang, Y. & Carmichael, G.C. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell Biol. 16, 1534–1542 (1996).
Hammell, C.M. et al. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell Biol. 22, 6441–6457 (2002).
Lei, E.P. & Silver, P.A. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 16, 2761–2766 (2002).
Rougemaille, M. et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 135, 308–321 (2008).
Daneholt, B. Assembly and transport of a premessenger RNP particle. Proc. Natl. Acad. Sci. USA 98, 7012–7017 (2001).
Lei, E.P., Krebber, H. & Silver, P.A. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15, 1771–1782 (2001).
Zenklusen, D., Vinciguerra, P., Wyss, J.C. & Stutz, F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell Biol. 22, 8241–8253 (2002).
Kiesler, E., Miralles, F. & Visa, N. HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr. Biol. 12, 859–862 (2002).
Hieronymus, H. & Silver, P.A. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33, 155–161 (2003).
Johnson, S.A., Cubberley, G. & Bentley, D.L. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell 33, 215–226 (2009).
Taniguchi, I. & Ohno, M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol. Cell Biol. 28, 601–608 (2008).
Abruzzi, K.C., Lacadie, S. & Rosbash, M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 23, 2620–2631 (2004).
Iglesias, N. et al. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 24, 1927–1938 (2010).
Köhler, A. & Hurt, E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8, 761–773 (2007).
Kim, M., Ahn, S.-H., Krogan, N.J., Greenblatt, J.F. & Buratowski, S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23, 354–364 (2004).
Xu, Z. et al. Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037 (2009).
Mandel, C.R., Bai, Y. & Tong, L. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65, 1099–1122 (2008).
Millevoi, S. & Vagner, S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 38, 2757–2774 (2010).
Zhao, J., Hyman, L. & Moore, C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445 (1999).
Guo, Z. & Sherman, F. 3′-end-forming signals of yeast messenger RNA. Trends Biochem. Sci. 21, 477–481 (1996).
Graber, J.H., McAllister, G.D. & Smith, T.F. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30, 1851–1858 (2002).
Gross, S. & Moore, C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. USA 98, 6080–6085 (2001).
Leeper, T.C., Qu, X., Lu, C., Moore, C. & Varani, G. Novel protein–protein contacts facilitate mRNA 3′-processing signal recognition by Rna15 and Hrp1. J. Mol. Biol. 401, 334–349 (2010).
Minvielle-Sebastia, L. et al. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 17, 7454–7468 (1998).
Kim Guisbert, K.S., Li, H. & Guthrie, C. Alternative 3′ pre-mRNA processing in Saccharomyces cerevisiae is modulated by Nab4/Hrp1 in vivo. PLoS Biol. 5, e6 (2007).
Lykke-Andersen, S. & Jensen, T.H. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie 89, 1177–1182 (2007).
Grzechnik, P. & Kufel, J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell 32, 247–258 (2008).
Phatnani, H.P. & Greenleaf, A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 (2006).
Strässer, K. & Hurt, E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413, 648–652 (2001).
Saguez, C. et al. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol. Cell 31, 91–103 (2008).
Noble, C.G., Beuth, B. & Taylor, I.A. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 35, 87–99 (2007).
Saguez, C. & Jensen, T.H. Assembly of export-competent mRNP: it's all about being connected. Mol. Cell 33, 139–140 (2009).
Lutz, C.S. & Moreira, A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. WIREs RNA 2, 23–31 (2011).
Ji, Z. & Tian, B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4, e8419 (2009).
Mayr, C. & Bartel, D.P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009).
Ozsolak, F. et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143, 1018–1029 (2010).
Preker, P.J., Kim, K.S. & Guthrie, C. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8, 969–980 (2002).
Kim, H. et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 17, 1279–1286 (2010).
Mayer, A. et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17, 1272–1278 (2010).
Mandart, E. & Parker, R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol. Cell Biol. 15, 6979–6986 (1995).
Mandart, E. Effects of mutations in the Saccharomyces cerevisiae RNA14 gene on the abundance and polyadenylation of its transcripts. Mol. Gen. Genet. 258, 16–25 (1998).
Kessler, M.M. et al. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 11, 2545–2556 (1997).
Gross, S. & Moore, C.L. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell Biol. 21, 8045–8055 (2001).
Bucheli, M.E., He, X., Kaplan, C.D., Moore, C.L. & Buratowski, S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA 13, 1756–1764 (2007).
Veraldi, K.L. et al. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell Biol. 21, 1228–1238 (2001).
Gawande, B., Robida, M.D., Rahn, A. & Singh, R. Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J. 25, 1263–1272 (2006).
Bruhn, L., Munnerlyn, A. & Grosschedl, R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 11, 640–653 (1997).
Virbasius, C.M., Wagner, S. & Green, M.R. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell 4, 219–228 (1999).
de Vries, H. et al. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 19, 5895–5904 (2000).
Takagaki, Y., Seipelt, R.L., Peterson, M.L. & Manley, J.L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87, 941–952 (1996).
Kaida, D. et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468, 664–668 (2010).
Stutz, F. et al. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6, 638–650 (2000).
Zhao, J., Kessler, M., Helmling, S., O'Connor, J.P. & Moore, C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell Biol. 19, 7733–7740 (1999).
Mangone, M. et al. The Landscape of C. elegans 3′UTRs. Science 329, 432–435 (2010).
Acknowledgements
This work was supported by US National Institutes of Health grant GM58613 to D.B. We thank C. Moore (Tufts University) for GAL7 plasmids and advice on processing extracts, R. Zhao (University of Colorado) for recombinant Sub2, C. Wang and M. Covarrubias (City of Hope Functional Genomics Core) for array hybridization, J. Dover and J. Castoe for Illumina sequencing, and T. Blumenthal and J. Hesselberth (University of Colorado) for helpful comments.
Author information
Authors and Affiliations
Contributions
S.A.J., B.E. and D.L.B. designed and conducted the experiments. H.K. wrote the software and carried out the informatics. S.A.J., H.K. and D.L.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Table 1 and Supplementary Methods (PDF 6697 kb)
Supplementary Data 1
Gene lists used in this study (XLS 153 kb)
Rights and permissions
About this article
Cite this article
Johnson, S., Kim, H., Erickson, B. et al. The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol 18, 1164–1171 (2011). https://doi.org/10.1038/nsmb.2126
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2126
This article is cited by
-
The functional complexity of the RNA-binding protein Yra1: mRNA biogenesis, genome stability and DSB repair
Current Genetics (2020)
-
SAGA DUBm-mediated surveillance regulates prompt export of stress-inducible transcripts for proteostasis
Nature Communications (2019)
-
Coupling mRNA processing with transcription in time and space
Nature Reviews Genetics (2014)
-
The regulatory role of Pcf11-similar-4 (PCFS4) in Arabidopsis development by genome-wide physical interactions with target loci
BMC Genomics (2013)
-
Chtop is a component of the dynamic TREX mRNA export complex
The EMBO Journal (2013)