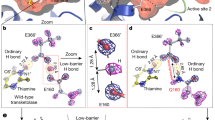
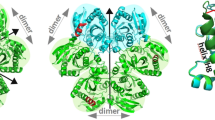
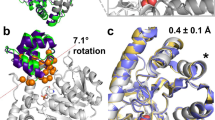
Abstract
Allostery has been studied for many decades, yet it remains challenging to determine experimentally how it occurs at a molecular level. We have developed an approach combining isothermal titration calorimetry, circular dichroism and nuclear magnetic resonance spectroscopy to quantify allostery in terms of protein thermodynamics, structure and dynamics. This strategy was applied to study the interaction between aminoglycoside N-(6′)-acetyltransferase-Ii and one of its substrates, acetyl coenzyme A. It was found that homotropic allostery between the two active sites of the homodimeric enzyme is modulated by opposing mechanisms. One follows a classical Koshland-Némethy-Filmer (KNF) paradigm, whereas the other follows a recently proposed mechanism in which partial unfolding of the subunits is coupled to ligand binding. Competition between folding, binding and conformational changes represents a new way to govern energetic communication between binding sites.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Perutz, M.F. Mechanisms of cooperativity and allosteric regulation in proteins. Q. Rev. Biophys. 22, 139–237 (1989).
Monod, J., Wyman, J. & Changeux, J.P. On nature of allosteric transitions—a plausible model. J. Mol. Biol. 12, 88–118 (1965).
Fenton, A.W. Allostery: an illustrated definition for the 'second secret of life'. Trends Biochem. Sci. 33, 420–425 (2008).
DeDecker, B.S. Allosteric drugs: thinking outside the active-site box. Chem. Biol. 7, R103–R107 (2000).
Koshland, D.E., Nemethy, G. & Filmer, D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385 (1966).
Perutz, M.F., Wilkinson, A.J., Paoli, M. & Dodson, G.G. The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu. Rev. Biophys. Biomol. Struct. 27, 1–34 (1998).
Helmstaedt, K., Krappmann, S. & Braus, G.H. Allosteric regulation of catalytic activity: Escherichia coli aspartate transcarbamoylase versus yeast chorismate mutase. Microbiol. Mol. Biol. Rev. 65, 404–421 (2001).
Cui, Q. & Karplus, M. Allostery and cooperativity revisited. Protein Sci. 17, 1295–1307 (2008).
Kern, D. & Zuiderweg, E.R.P. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 13, 748–757 (2003).
Hilser, V.J. & Thompson, E.B. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl. Acad. Sci. USA 104, 8311–8315 (2007).
Reichheld, S.E., Yu, Z. & Davidson, A.R. The induction of folding cooperativity by ligand binding drives the allosteric response of tetracycline repressor. Proc. Natl. Acad. Sci. USA 106, 22263–22268 (2009).
Popovych, N., Sun, S.J., Ebright, R.H. & Kalodimos, C.G. Dynamically driven protein allostery. Nat. Struct. Mol. Biol. 13, 831–838 (2006).
Cooper, A. & Dryden, D.T.F. Allostery without conformational change—a plausible model. Eur. Biophys. J. 11, 103–109 (1984).
Tzeng, S.R. & Kalodimos, C.G. Dynamic activation of an allosteric regulatory protein. Nature 462, 368–372 (2009).
Gandhi, P.S., Chen, Z.W., Mathews, F.S. & Di Cera, E. Structural identification of the pathway of long-range communication in an allosteric enzyme. Proc. Natl. Acad. Sci. USA 105, 1832–1837 (2008).
Lockless, S.W. & Ranganathan, R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295–299 (1999).
Tsai, C.J., Kumar, S., Ma, B.Y. & Nussinov, R. Folding funnels, binding funnels, and protein function. Protein Sci. 8, 1181–1190 (1999).
Tsai, C.J., Ma, B.Y. & Nussinov, R. Folding and binding cascades: shifts in energy landscapes. Proc. Natl. Acad. Sci. USA 96, 9970–9972 (1999).
Kumar, S., Ma, B.Y., Tsai, C.J., Sinha, N. & Nussinov, R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 9, 10–19 (2000).
Pan, H., Lee, J.C. & Hilser, V.J. Binding sites in Escherichia coli dihydrofolate reductase communicate by modulating the conformational ensemble. Proc. Natl. Acad. Sci. USA 97, 12020–12025 (2000).
Volkman, B.F., Lipson, D., Wemmer, D.E. & Kern, D. Two-state allosteric behavior in a single-domain signaling protein. Science 291, 2429–2433 (2001).
Gunasekaran, K., Ma, B.Y. & Nussinov, R. Is allostery an intrinsic property of all dynamic proteins? Proteins 57, 433–443 (2004).
Clarkson, M.W., Gilmore, S.A., Edgell, M.H. & Lee, A.L. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry 45, 7693–7699 (2006).
Wright, G.D. & Ladak, P. Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob. Agents Chemother. 41, 956–960 (1997).
Burk, D.L., Xiong, B., Breitbach, C. & Berghuis, A.M. Structures of aminoglycoside acetyltransferase AAC(6′)-Ii in a novel crystal form: structural and normal-mode analyses. Acta Crystallogr. D Biol. Crystallogr. 61, 1273–1279 (2005).
Wybenga-Groot, L.E., Draker, K.-a., Wright, G.D. & Berghuis, A.M. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure 7, 497–507 (1999).
Burk, D.L., Ghuman, N., Wybenga-Groot, L.E. & Berghuis, A.M. X-ray structure of the AAC(6′)-Ii antibiotic resistance enzyme at 1.8 angstrom resolution; examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 12, 426–437 (2003).
Gao, F., Yan, X.X., Baettig, O.M., Berghuis, A.M. & Auclair, K. Regio- and chemoselective 6′-N-derivatization of aminoglycosides: bisubstrate inhibitors as probes to study aminoglycoside 6′-N-acetyltransferases. Angew. Chem. Int. Edn Engl. 44, 6859–6862 (2005).
Freiburger, L.A., Auclair, K. & Mittermaier, A.K. Elucidating protein binding mechanisms by variable-c ITC. ChemBioChem 10, 2871–2873 (2009).
Fisher, H.F., Colen, A.H. & Medary, R.T. Temperature-dependent ΔC p0 generated by a shift in equilibrium between macrostates of an enzyme. Nature 292, 271–272 (1981).
Cliff, M.J., Williams, M.A., Brooke-Smith, J., Barford, D. & Ladbury, J.E. Molecular recognition via coupled folding and binding in a TPR domain. J. Mol. Biol. 346, 717–732 (2005).
Keramisanou, D. et al. Disorder-order folding transitions underlie catalysis in the helicase motor of SecA. Nat. Struct. Mol. Biol. 13, 594–602 (2006).
Adler, A.J., Greenfield, N.J. & Fasman, G.D. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 27, 675–735 (1973).
Saxena, V.P. & Wetlaufer, D.B. A New Basis for interpreting circular dichroic spectra of proteins. Proc. Natl. Acad. Sci. USA 68, 969–972 (1971).
Uversky, V.N. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11, 739–756 (2002).
Farrow, N.A., Zhang, O., Forman-Kay, J.D. & Kay, L.E. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4, 727–734 (1994).
Farmer, B.T. Localizing the NADP(+) binding site on the MurB enzyme by NMR. Nat. Struct. Biol. 3, 995–997 (1996).
Koshland, D.E. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA 44, 98–104 (1958).
Foote, J. & Milstein, C. Conformational isomerism and the diversity of antibodies. Proc. Natl. Acad. Sci. USA 91, 10370–10374 (1994).
Boehr, D.D., Nussinov, R. & Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009).
Koshland, D.E. & Hamadani, K. Proteomics and models for enzyme cooperativity. J. Biol. Chem. 277, 46841–46844 (2002).
Bosshard, H.R. Molecular recognition by induced fit: how fit is the concept? News Physiol. Sci. 16, 171–173 (2001).
Hammes, G.G., Chang, Y.C. & Oas, T.G. Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. USA 106, 13737–13741 (2009).
Weikl, T.R. & von Deuster, C. Selected-fit versus induced-fit protein binding: kinetic differences and mutational analysis. Proteins 75, 104–110 (2009).
Radley, T.L., Markowska, A.I., Bettinger, B.T., Ha, J.H. & Loh, S.N. Allosteric switching by mutually exclusive folding of protein domains. J. Mol. Biol. 332, 529–536 (2003).
Laine, O., Streaker, E.D., Nabavi, M., Fenselau, C.C. & Beckett, D. Allosteric signaling in the biotin repressor occurs via local folding coupled to global dampening of protein dynamics. J. Mol. Biol. 381, 89–101 (2008).
Ehlert, F.J. Estimation of the affinities of allosteric ligands using radioligand binding and pharmacological null methods. Mol. Pharmacol. 33, 187–194 (1988).
Marley, J., Lu, M. & Bracken, C. A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 (2001).
Cavanagh, J., Fairbrother, W., Palmer, A. & Skelton, N. Protein NMR Spectroscopy Principles and Practice (Academic Press, Inc., 1996).
Kay, L.E., Keifer, P. & Saarinen, T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665 (1992).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Acknowledgements
The authors would like to thank G.D. Wright (McMaster University) for providing the AAC(6′)-Ii expression construct, as well as C.E. Ang and K. Shopsowitz for their assistance with data collection and analysis. This research was funded by operating grants from the Canadian Institutes of Health Research (CIHR) (K.A., A.K.M. and A.M.B.). L.A.F. and O.M.B. were supported through a CIHR training grant and CIHR scholarship, respectively. A.M.B. holds a Canada Research Chair in Structural Biology. A.M.B. and A.K.M. are supported by the Groupe de Recherche Axé sur la Structure des Protéines (GRASP). NMR experiments were recorded at the Québec/Eastern Canada High Field NMR Facility, supported by the Natural Sciences and Engineering Research Council of Canada, the Canada Foundation for Innovation, the Ministère de la Recherche, de la Science et de la Technologie du Québec, and McGill University.
Author information
Authors and Affiliations
Contributions
L.A.F. and O.M.B. collected the data. L.A.F., O.M.B. and T.S. analyzed the data. A.K.M., K.A. and A.M.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1 and Supplementary Methods (PDF 799 kb)
Rights and permissions
About this article
Cite this article
Freiburger, L., Baettig, O., Sprules, T. et al. Competing allosteric mechanisms modulate substrate binding in a dimeric enzyme. Nat Struct Mol Biol 18, 288–294 (2011). https://doi.org/10.1038/nsmb.1978
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1978
This article is cited by
-
Isothermal titration calorimetry
Nature Reviews Methods Primers (2023)
-
Backbone resonance assignment of an insect arylalkylamine N-acetyltransferase from Bombyx mori reveals conformational heterogeneity
Biomolecular NMR Assignments (2017)
-
Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions
Nature Protocols (2016)
-
Large interdomain rearrangement triggered by suppression of micro- to millisecond dynamics in bacterial Enzyme I
Nature Communications (2015)
-
Disordered allostery: lessons from glucocorticoid receptor
Biophysical Reviews (2015)