Abstract
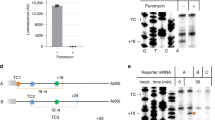
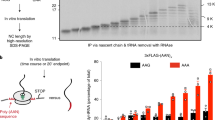
Nonsense-mediated decay (NMD) degrades mRNA containing premature translation termination codons. In yeast, NMD substrates are decapped and digested exonucleolytically from the 5′ end. Despite the requirement for translation in recognition, degradation of nonsense-containing mRNA is considered to occur in ribosome-free cytoplasmic P bodies. We show decapped nonsense-containing mRNA associate with polyribosomes, indicating that recognition and degradation are tightly coupled and that polyribosomes are major sites for degradation of aberrant mRNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Isken, O. & Maquat, L.E. Genes Dev. 21, 1833–1856 (2007).
Behm-Ansmant, I. et al. FEBS Lett. 581, 2845–2853 (2007).
Amrani, N., Sachs, M.S. & Jacobson, A. Nat. Rev. Mol. Cell Biol. 7, 415–425 (2006).
Baker, K.E. & Parker, R. Curr. Opin. Cell Biol. 16, 293–299 (2004).
Muhlrad, D. & Parker, R. Nature 370, 578–581 (1994).
Lejeune, F., Li, X. & Maquat, L.E. Mol. Cell 12, 675–687 (2003).
Eberle, A.B., Lykke-Andersen, S., Muhlemann, O. & Jensen, T.H. Nat. Struct. Mol. Biol. 16, 49–55 (2009).
Atkin, A.L. et al. J. Biol. Chem. 272, 22163–22172 (1997).
Mangus, D.A. & Jacobson, A. Methods 17, 28–37 (1999).
Sheth, U. & Parker, R. Cell 125, 1095–1109 (2006).
Isken, O. et al. Cell 133, 314–327 (2008).
Muhlrad, D. & Parker, R. Mol. Biol. Cell 10, 3971–3978 (1999).
Hu, W., Sweet, T.J., Chamnongpol, S., Baker, K.E. & Coller, J. Nature 461, 225–229 (2009).
Stalder, L. & Muhlemann, O. RNA 15, 1265–1273 (2009).
Kshirsagar, M. & Parker, R. Genetics 166, 729–739 (2004).
Decker, C.J., Teixeira, D. & Parker, R. J. Cell Biol. 179, 437–449 (2007).
Rehwinkel, J., Behm-Ansmant, I., Gatfield, D. & Izaurralde, E. RNA 11, 1640–1647 (2005).
Coller, J. & Parker, R. Cell 122, 875–886 (2005).
Inada, T. et al. RNA 8, 948–958 (2002).
Wang, Z., Jiao, X., Carr-Schmid, A. & Kiledjian, M. Proc. Natl. Acad. Sci. USA 99, 12663–12668 (2002).
Acknowledgements
The authors thank members of the Baker and Coller laboratories for critical evaluation of the manuscript. W.H. is supported by the American Heart Association. C.P. is supported by an undergraduate stipend provided by the US National Science Foundation. Funding was provided by the US National Institutes of Health (GM080465).
Author information
Authors and Affiliations
Contributions
W.H., J.C. and K.E.B. designed experimental strategies; W.H. and C.P. performed experimental analysis and interpreted data; J.C. and K.E.B. interpreted data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplenentary Figures 1–4, Supplementary Table 1 and Supplementary Data (PDF 3446 kb)
Rights and permissions
About this article
Cite this article
Hu, W., Petzold, C., Coller, J. et al. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol 17, 244–247 (2010). https://doi.org/10.1038/nsmb.1734
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1734
This article is cited by
-
Quality and quantity control of gene expression by nonsense-mediated mRNA decay
Nature Reviews Molecular Cell Biology (2019)
-
Molecular autopsy provides evidence for widespread ribosome-phased mRNA fragmentation
Nature Structural & Molecular Biology (2018)
-
Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA
The EMBO Journal (2013)
-
Loss of nonsense mediated decay suppresses mutations in Saccharomyces cerevisiae TRA1
BMC Genetics (2012)