Abstract
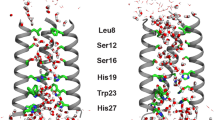
Influenza B virus contains an integral membrane protein, BM2, that oligomerizes in the viral membrane to form a pH-activated proton channel. Here we report the solution structures of both the membrane-embedded channel domain and the cytoplasmic domain of BM2. The channel domain assumes a left-handed coiled-coil tetramer formation with a helical packing angle of −37° to form a polar pore in the membrane for conducting ions. Mutagenesis and proton flux experiments identified residues involved in proton relay and suggest a mechanism of proton conductance. The cytoplasmic domain of BM2 also forms a coiled-coil tetramer. It has a bipolar charge distribution, in which a negatively charged region interacts specifically with the M1 matrix protein that is involved in packaging the genome in the virion. This interaction suggests BM2 also recruits matrix proteins to the cell surface during virus budding, making BM2 an unusual membrane protein with the dual roles of conducting ions and recruiting proteins to the membrane.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hatta, M., Goto, H. & Kawaoka, Y. Influenza B virus requires BM2 protein for replication. J. Virol. 78, 5576–5583 (2004).
Pinto, L.H. & Lamb, R.A. The M2 proton channels of influenza A and B viruses. J. Biol. Chem. 281, 8997–9000 (2006).
Hay, A.J., Wolstenholme, A.J., Skehel, J.J. & Smith, M.H. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4, 3021–3024 (1985).
Helenius, A. Unpacking the incoming influenza virus. Cell 69, 577–578 (1992).
Paterson, R.G., Takeda, M., Ohigashi, Y., Pinto, L.H. & Lamb, R.A. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology 306, 7–17 (2003).
Mould, J.A. et al. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell 5, 175–184 (2003).
Pinto, L.H. et al. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc. Natl. Acad. Sci. USA 94, 11301–11306 (1997).
Kovacs, F.A., Denny, J.K., Song, Z., Quine, J.R. & Cross, T.A. Helix tilt of the M2 transmembrane peptide from influenza A virus: an intrinsic property. J. Mol. Biol. 295, 117–125 (2000).
Kukol, A., Adams, P.D., Rice, L.M., Brunger, A.T. & Arkin, T.I. Experimentally based orientational refinement of membrane protein models: A structure for the influenza A M2 H+ channel. J. Mol. Biol. 286, 951–962 (1999).
Schnell, J.R. & Chou, J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451, 591–595 (2008).
Wang, J., Kim, S., Kovacs, F. & Cross, T.A. Structure of the transmembrane region of the M2 protein H+ channel. Protein Sci. 10, 2241–2250 (2001).
Stouffer, A.L. et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 451, 596–599 (2008).
Ma, C. et al. Identification of the pore-lining residues of the BM2 ion channel protein of influenza B virus. J. Biol. Chem. 283, 15921–15931 (2008).
Chen, B.J., Leser, G.P., Jackson, D. & Lamb, R.A. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82, 10059–10070 (2008).
McCown, M.F. & Pekosz, A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 80, 8178–8189 (2006).
Imai, M., Kawasaki, K. & Odagiri, T. Cytoplasmic domain of influenza B virus BM2 protein plays critical roles in production of infectious virus. J. Virol. 82, 728–739 (2008).
Imai, M., Watanabe, S., Ninomiya, A., Obuchi, M. & Odagiri, T. Influenza B virus BM2 protein is a crucial component for incorporation of viral ribonucleoprotein complex into virions during virus assembly. J. Virol. 78, 11007–11015 (2004).
Pielak, R.M., Schnell, J.R. & Chou, J.J. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc. Natl. Acad. Sci. USA 106, 7379–7384 (2009).
Xu, C. et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 135, 702–713 (2008).
Harbury, P.B., Zhang, T., Kim, P.S. & Alber, T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262, 1401–1407 (1993).
Felder, C., Prilusky, J., Silman, I. & Sussman, J. A server and database for dipole moments of proteins. Nucleic Acids Res. 35 (special Web Servers Issue) (2007).
Sha, B. & Luo, M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat. Struct. Biol. 4, 239–244 (1997).
Otomo, K., Toyama, A., Miura, T. & Takeuchi, H. Interactions between histidine and tryptophan residues in the BM2 proton channel from influenza B virus. J. Biochem. 145, 543–554 (2009).
Du, Q.S., Huang, R.B., Wang, C.H., Li, X.M. & Chou, K.C. Energetic analysis of the two controversial drug binding sites of the M2 proton channel in influenza A virus. J. Theor. Biol. 259, 159–164 (2009).
Chou, J.J., Matsuo, H., Duan, H. & Wagner, G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell 94, 171–180 (1998).
Smart, O.S., Goodfellow, J.M. & Wallace, B.A. The pore dimensions of gramicidin A. Biophys. J. 65, 2455–2460 (1993).
Bax, A. et al. Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol. 239, 79–105 (1994).
Lorieau, J., Yao, L. & Bax, A. Liquid crystalline phase of G-tetrad DNA for NMR study of detergent-solubilized proteins. J. Am. Chem. Soc. 130, 7536–7537 (2008).
Schwieters, C.D., Kuszewski, J., Tjandra, N. & Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 66–74 (2002).
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999).
Acknowledgements
We thank M. Berardi for insightful discussion on the interaction between BM2 and M1 matrix protein. This work was supported by grants from the US National Institutes of Health (AI054520 to J.J.C.) and the Pew Scholars Program in Biomedical Sciences to J.J.C.
Author information
Authors and Affiliations
Contributions
J.W. and J.J.C. determined the BM2 structures; R.M.P. performed proton conductance assays; J.W. and M.A.M. performed the BM2-BM1 interaction experiment; J.J.C., R.M.P. and J.W. wrote the paper; J.J.C. supervised the research.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Figures 1–13 (PDF 1363 kb)
Rights and permissions
About this article
Cite this article
Wang, J., Pielak, R., McClintock, M. et al. Solution structure and functional analysis of the influenza B proton channel. Nat Struct Mol Biol 16, 1267–1271 (2009). https://doi.org/10.1038/nsmb.1707
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1707
This article is cited by
-
Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism
Nature Structural & Molecular Biology (2020)
-
Progresses in Predicting Post-translational Modification
International Journal of Peptide Research and Therapeutics (2020)
-
M2e-based universal influenza vaccines: a historical overview and new approaches to development
Journal of Biomedical Science (2019)
-
Structure determination protocol for transmembrane domain oligomers
Nature Protocols (2019)
-
The Transmembrane Conformation of the Influenza B Virus M2 Protein in Lipid Bilayers
Scientific Reports (2019)