Abstract
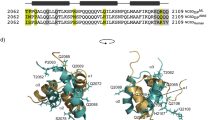
Nuclear receptors are transcription factors that activate gene expression in response to ligands. The C-terminal helix (helix 12) of the ligand-binding domain plays a critical role in the activation mechanism. When bound to activating ligands, helix 12 adopts a conformation that promotes the binding of co-activator proteins. Helix 12 also adopts this 'active' position in several ligand-free structures, raising questions as to the exact role of helix 12. We proposed that the dynamic properties of helix 12 may be critical for the activation mechanism and, to test this, have used fluorescence anisotropy techniques to directly monitor the mobility of helix 12 in PPARγ. Our results suggest that helix 12 is significantly more mobile than the main body of the protein. Upon ligand binding, helix 12 shows reduced mobility, accounting for its role as a molecular switch. We also show that natural mutations in human PPARγ, associated with severe insulin resistance and diabetes mellitus, exhibit perturbations in the dynamic behavior of helix 12. Our findings provide the first direct observations of the mobility of helix 12 and suggest that the dynamic properties of this helix are key to the regulation of transcriptional activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Darimont, B.D. et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12, 3343–3356 (1998).
Shiau, A.K. et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937 (1998).
Nolte, R. et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor. Nature 395, 137–143 (1998).
Bourguet, W., Ruff, M., Chambon, P., Gronemeyer, H. & Moras, D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature 375, 377–382 (1995).
Brzozowski, A.M. et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389, 753–758 (1997).
Pike, A.C. et al. Structure of the ligand-binding domain of oestrogen receptor βε in the presence of a partial agonist and a full antagonist. EMBO J. 18, 4608–4618 (1999).
Gampe, R.T. Jr. et al. Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 14, 2229–2241 (2000).
Bourguet, W. et al. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5, 289–298 (2000).
Pike, A.C. et al. Structural insights into the mode of action of a pure antiestrogen. Structure 9, 145–153 (2001).
Watkins, R.E. et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292, 2329–2333 (2001).
Clayton, G.M., Peak-Chew, S.Y., Evans, R.M. & Schwabe, J.W. The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation. Proc. Natl. Acad. Sci. USA 98, 1549–1554 (2001).
Xu, H.E. et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα. Nature 415, 813–817 (2002).
Hu, X. & Lazar, M.A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402, 93–96 (1999).
Perissi, V. et al. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 13, 3198–3208 (1999).
Nagy, L. et al. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 13, 3209–3216 (1999).
Webb, P. et al. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs). Mol. Endocrinol. 14, 1976–1985 (2000).
Barroso, I. et al. Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402, 880–883 (1999).
Chong, S. et al. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192, 271–281 (1997).
Lehmann, J.M. et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ). J. Biol. Chem. 270, 12953–12956 (1995).
Yguerabide, J., Epstein, H.F. & Stryer, L. Segmental flexibility in an antibody molecule. J. Mol. Biol. 51, 573–590 (1970).
Vergani, B. et al. Backbone dynamics of Tet repressor α8–α9 loop. Biochemistry 39, 2759–2768 (2000).
Gangal, M. et al. Backbone flexibility of five sites on the catalytic subunit of cAMP-dependent protein kinase in the open and closed conformations. Biochemistry 37, 13728–13735 (1998).
Mielke, T., Alexiev, U., Glasel, M., Otto, H. & Heyn, M.P. Light-induced changes in the structure and accessibility of the cytoplasmic loops of rhodopsin in the activated MII state. Biochemistry 41, 7875–7884 (2002).
Clore, G.M., Driscoll, P.C., Wingfield, P.T. & Gronenborn, A.M. Analysis of the backbone dynamics of interleukin-1β using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry 29, 7387–7401 (1990).
Johnson, B.A. et al. Ligand-induced stabilization of PPARγ monitored by NMR spectroscopy: implications for nuclear receptor activation. J. Mol. Biol. 298, 187–194 (2000).
Cronet, P. et al. Structure of the PPARα and -γ ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure 9, 699–706 (2001).
Gangloff, M. et al. Crystal structure of a mutant hERα ligand-binding domain reveals key structural features for the mechanism of partial agonism. J. Biol. Chem. 276, 15059–15065 (2001).
Schulman, I.G., Juguilon, H. & Evans, R.M. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol. Cell. Biol. 16, 3807–3813 (1996).
Tanenbaum, D.M., Wang, Y., Williams, S.P. & Sigler, P.B. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc. Natl. Acad. Sci. USA 95, 5998–6003 (1998).
Love, J.D. et al. The structural basis for the specificity of retinoid-X-receptor selective agonists: new insights into the role of helix H12. J. Biol. Chem. 277, 11385–11391 (2002).
Gregory, C., Hayes, M., Jones, G. & Pantos, E. FLUOR — a program to analyse fluorescence data. in Daresbury Laboratory Technical Memorandum (Daresbury Laboratory, Warrington; 1994).
Brown, K.K. et al. A novel N-aryl tyrosine activator of peroxisome proliferator-activated receptor-γ reverses the diabetic phenotype of the Zucker diabetic fatty rat. Diabetes 48, 1415–1424 (1999).
Li, F. et al. Evidence for an internal entropy contribution to phosphoryl transfer: a study of domain closure, backbone flexibility, and the catalytic cycle of cAMP-dependent protein kinase. J. Mol. Biol. 315, 459–469 (2002).
Acknowledgements
We are grateful to D. Veprintsev for assistance with the steady state measurements, T. Willson and Glaxo-SmithKline for the gift of PPARγ ligands, D. Owen for synthesis of fluorophore and co-activator peptide, D. Shaw and M. Martin-Fernandez for assistance at the SRS beamline and T. Mielke, P. Evans, D. Neuhaus and D. Rhodes for helpful discussions. The work was supported in part by HFSP and EU-RTN grants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kallenberger, B., Love, J., Chatterjee, V. et al. A dynamic mechanism of nuclear receptor activation and its perturbation in a human disease. Nat Struct Mol Biol 10, 136–140 (2003). https://doi.org/10.1038/nsb892
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb892
This article is cited by
-
Definition of functionally and structurally distinct repressive states in the nuclear receptor PPARγ
Nature Communications (2019)
-
Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPARγ
Scientific Reports (2018)
-
Integrated in silico–in vitro screening of ovarian cancer peroxisome proliferator-activated receptor-γ agonists against a biogenic compound library
Medicinal Chemistry Research (2018)
-
Structures of PPARγ complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Scientific Reports (2017)
-
Allosteric Pathways in the PPARγ-RXRα nuclear receptor complex
Scientific Reports (2016)