Abstract
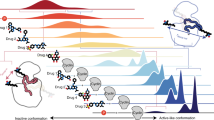
Cyclin from herpesvirus saimiri (Vcyclin) preferentially forms complexes with cyclin-dependent kinase 6 (CDK6) from primate host cells. These complexes show higher kinase activity than host cell CDKs in complex with cellular cyclins and are resistant to cyclin-dependent inhibitory proteins (CDKIs). The crystal structure of human CDK6–Vcyclin in an active state was determined to 3.1 Å resolution to better understand the structural basis of CDK6 activation by viral cyclins. The unphosphorylated CDK6 in complex with Vcyclin has many features characteristic of cyclinA-activated, phosphorylated CDK2. There are, however, differences in the conformation at the tip of the T-loop and its interactions with Vcyclin. Residues in the N-terminal extension of Vcyclin wrap around the tip of the CDK6 T-loop and form a short β-sheet with the T-loop backbone. These interactions lead to a 20% larger buried surface in the CDK6–Vcyclin interface than in the CDK2–cyclinA complex and are probably largely responsible for the specificity of Vcyclin for CDK6 and resistance of the complex to inhibition by INK-type CDKIs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Sherr, C.J. Cell 73, 1059–1065 (1993).
Sherr, C.J. Cell 79, 551–555 (1994).
Pavletich, N.P. J. Mol. Biol. 287, 821–828 (1999).
Hall, M. & Peters, G. Adv. Cancer Res. 68, 67–108 (1996).
Sherr, C.J. Science 274, 1672–1677 (1996).
Mittnacht, S. & Boshoff, C. Rev. Med. Virol. 10, 175–184 (2000).
Jung, J.U., Stager, M. & Desrosiers, R.C. Mol. Cell. Biol. 14, 7235–7244 (1994).
Li, M. et al. J. Virol. 71, 1984–1991 (1997).
Godden-Kent, D. et al. J. Virol. 71, 4193–4198 (1997).
Swanton, C. et al. Nature 390, 184–187 (1997).
DeBondt, H.L. et al. Nature 363, 595–602 (1993).
Jeffrey, D.P. et al. Nature 376, 313–320 (1995).
Schulze-Gahmen, U., Jung, J.U. & Kim, S.-H. Structure 7, 245–254 (1999).
Russo, A.A., Jeffrey, P.D. & Pavletich, N.P. Nature Struct. Biol. 3, 696–700 (1996).
Card, G.L., Knowles, P., Laman, H., Jones, N. & McDonald, N.Q. EMBO J. 19, 2877–2888 (2000).
Jeffrey, P.D., Tong, L. & Pavletich, N.P. Genes Dev. 14, 3115–3125 (2000).
Russo, A.A., Tong, L., Lee, J.-O., Jeffrey, P.D. & Pavletich, N.P. Nature 395, 237–243 (1998).
Brotherton, D.H. et al. Nature 395, 244–250 (1998).
Brown, N.R., Nobel, M.E.M., Endicott, J.A. & Johnson, L.N. Nature Cell Biol. 1, 438–443 (1999).
Hol, W.G.J., Van Duijnen, P.T. & Berendsen, H.J.C. Nature 273, 443–446 (1978).
Kitagawa, M. et al. EMBO J. 15, 7060–7069 (1996).
Ellis, M. et al. EMBO J. 18, 644–653 (1999).
Russo, A.A., Jeffrey, P.D., Patten, A.K., Massague, J. & Pavletich, N.P. Nature 382, 325–331 (1996).
Schulze-Gahmen, U. & Kim, S.-H. Acta Crystallogr. D 57, 1287–1289 (2001).
Otwinski, Z.M., & Minor, W. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4 Acta Crystallogr. D 50, 760–763 (1994).
Brünger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjelgaard, M. Acta Crystallogr. D 47, 110–119 (1991).
Rossman, M.G. & Argos, P.A. J. Biol. Chem. 250, 7525–7532 (1975).
Sheriff, S., Hendrickson, W.A. & Smith, J.L. J. Mol. Biol. 197, 273–296 (1987).
Connolly, M.L. J. Appl. Crystallogr. 16, 548–558 (1983).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. Nucleic Acids Res. 22, 4673–4680 (1994).
Read, R.J. Acta Crystallogr. A 42, 140–149 (1986).
Acknowledgements
We thank J. Brandsen for his exploratory work on protein expression and purification and E. Berry for helpful discussions. This work was supported by grants from NIH and DOE.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schulze-Gahmen, U., Kim, SH. Structural basis for CDK6 activation by a virus-encoded cyclin. Nat Struct Mol Biol 9, 177–181 (2002). https://doi.org/10.1038/nsb756
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb756