Abstract
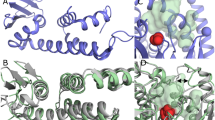
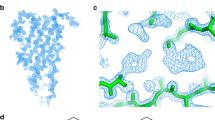
Eps15 homology (EH) domains are protein interaction modules that recognize Asn-Pro-Phe (NPF) motifs in their biological ligands to mediate critical events during endocytosis and signal transduction. To elucidate the structural basis of the EH–NPF interaction, the solution structures of two EH–NPF complexes were solved using NMR spectroscopy. The first complex contains a peptide representing the Hrb C-terminal NPFL motif; the second contains a peptide in which an Arg residue substitutes the C-terminal Leu. The NPF residues are almost completely embedded in a hydrophobic pocket on the EH domain surface and the backbone of NPFX adopts a conformation reminiscent of the Asx-Pro type I β-turn motif. The residue directly following NPF is crucial for recognition and is required to complete the β-turn. Five amino acids on the EH surface mediate specific recognition of this residue through hydrophobic and electrostatic contacts. The complexes explain the selectivity of the second EH domain of Eps15 for NPF over DPF motifs and reveal a critical aromatic interaction that provides a conserved anchor for the recognition of FW, WW, SWG and HTF ligands by other EH domains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Santolini, E., Salcini, A.E., Kay, B.K., Yamabhai, M. & Di Fiore, P.P. Exp. Cell Res. 253 , 186–209 (1999).
Yamabhai, M., et al. J. Biol. Chem. 273, 31401– 31407 (1998).
Chen, H. et al. Nature 394, 793–797 (1998).
Haffner, C. et al. FEBS Lett. 419, 175– 180 (1997).
Wendland, B. & Emr, S.D. J. Cell Biol. 141, 71–84 (1998).
Salcini, A.E. et al. Genes Dev. 11, 2239– 2249 (1997).
Doria, M. et al. J. Cell Biol. 147, 1379– 1384 (1999).
de Beer, T., Carter, R.E., Lobel-Rice, K.E., Sorkin, A. & Overduin, M. Science 281, 1357–1360 (1998).
Whitehead, B., Tessari, M., Carotenuto, A., van Bergen en Henegouwen, P.M. & Vuister, G.W. Biochemistry 38, 11271–11277 ( 1999).
Enmon, J.L., de Beer, T. & Overduin, M. Biochemistry 39, 4309– 4319 (2000).
Paoluzi, S. et al. EMBO J. 17, 6541–6550 (1998).
Wilson, D.R. & Finlay, B.B. Protein Eng. 10, 519–529 (1997).
Cupers, P., ter Haar, E., Boll, W. & Kirchhausen, T. J. Biol. Chem. 272, 33430–33434 ( 1997).
Nakashima, S. et al. EMBO J. 18, 3629–3642 (1999).
Fazioli, F., Minichiello, L., Matoskova, B., Wong, W.T. & Di Fiore, P.P. Mol. Cell Biol. 13 , 5814–5828 (1993).
Aue, W.P., Bartholdi, E. & Ernst, R.R. J. Chem. Phys. 64, 2229– 2246 (1976).
Braunschweiler, L. & Ernst, R.R. J. Magn. Reson. 53, 521–528 ( 1983).
Griesinger, C., Otting, G., Wüthrich, K. & Ernst, R.R. J. Am. Chem. Soc. 110, 7870–7872 (1988).
Bax, A. & Davis, D.G. J. Magn. Reson. 63, 207–213 (1985).
Medvedeva, S., Simorre, J.-P., Brutscher, B., Guerlesquin, F. & Marion, D. FEBS Lett. 333, 251–256 (1993).
Bodenhausen, G. & Ruben, D.J. Chem. Phys. Lett. 69, 185–189 ( 1980).
Kay, L.E., Keifer, P. & Saarinen, T. J. Am. Chem. Soc. 114, 10663– 10665 (1992).
Jeener, J., Meier, H., Bachmann, P. & Ernst, R.R. J. Chem. Phys. 71, 4546–4553 ( 1979).
Zhang, O., Kay, L.E., Olivier, J.P. & Forman-Kay, J.D. J. Biomol. NMR 4, 845–858 (1994).
Delaglio, F. et al. J. Biomol. NMR 6, 277– 293 (1995).
Garrett, D.S, Powers, R., Gronenborn, A., and Clore, G.M. J. Magn Reson. 95, 214–220 (1991).
Fletcher, C.M., Jones, D.N.M., Diamond, R. & Neuhaus, D. J. Biomol. NMR 8, 292–310 (1996).
Brünger, A.T, X-PLOR, version 3.1. (Yale University Press, New Haven, Connecticut; 1992).
Laskowski, R.A., Rullman, J.A., MacArthur, M.W., Kaptein, R. & Thornton, J.M. J. Biomol. NMR 8, 477–486 ( 1996).
Humphrey, W.F., Dalke, A. & Schulten, K. J. Mol. Graphics 14, 33–38 (1996).
Acknowledgements
We thank J. Mamay for computational support and for software to manipulate VMD images and A. Sorkin, and D.N.M. Jones for insightful discussions. T.B. is a recipient of a National Institutes of Health NSRA Fellowship. A.N.H. is a recipient of an NIH training grant fellowship. Grant support to M.O. was provided by the National Institutes of Health and the Pew Scholar's program, and the NMR center is supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Beer, T., Hoofnagle, A., Enmon, J. et al. Molecular mechanism of NPF recognition by EH domains. Nat Struct Mol Biol 7, 1018–1022 (2000). https://doi.org/10.1038/80924
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/80924
This article is cited by
-
Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding
Nature Communications (2021)
-
A junctional PACSIN2/EHD4/MICAL-L1 complex coordinates VE-cadherin trafficking for endothelial migration and angiogenesis
Nature Communications (2021)
-
Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29
Nature Communications (2020)
-
Structural basis for the recognition of two consecutive mutually interacting DPF motifs by the SGIP1 μ homology domain
Scientific Reports (2016)
-
Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation
Nature Cell Biology (2015)