Abstract
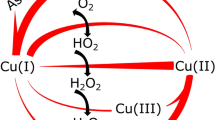
We have solved the structure of the Bacillus subtilis pectate lyase (BsPel) in complex with calcium. The structure consists of a parallel β-helix domain and a loop region. The αL-bounded β-strand seen in BsPel is a new element of protein structure and its frequent occurrence suggests it is an important characteristic of the parallel β-helix. A pronounced cleft is formed between the loops and the parallel β-helix domain and we propose that this is the active site cleft. Calcium, essential for the activity of the enzyme, binds at the bottom of this cleft and an arginine residue close to the calcium, which is conserved across all pectin and pectate lyases, may be involved in catalysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Pilnik, W. & Rombouts, F.M. In Enzymes and food processing (eds Birch,GG., Blakebrough,N. and Parker,K.J.), 105–128 (Applied Science Publishers Ltd., London; 1981.)
Yoder, M.D., Keen, N.T. and Jurnak,F. New domain motif: The structure of pectate lyase C, a plant virulence factor. Science 260, 1503–1507 (1993).
Yoder, M.D., Lietzke, S.E. & Jurnak, F. Unusual structural features in the parallel β-helix in pectate lyase. Structure 1, 241–245 (1993).
Nasser,W., Chalet, F. & Robert-Baudouy, J. Purification and characterization of extracellular pectate lyase from Bacillus subtilis. Biochemie 72, 689–695 (1990).
Nasser,W., Awadé, A.C., Reverchon, S. & Robert-Baudoy, J. Pectate lyase from Bacillus subtilis: molecular characterization of the gene, and properties of the cloned enzyme. FEBS Lett. 335, 319–326 (1993).
Bernstein, F.C. et al. The protein databank: a computer based archival file for macromolecular structures. J. molec. Biol. 112, 535–542 (1977).
Jones, T.A. & Thirup, S. Using known substructures in protein model-building and crystallography. EMBO J. 5, 819–822 (1986).
Carrell, C.J., Carrell, H.L., Erlebacher, J. & Glusker, J.P. Structural aspects of metal ion-carboxylate interactions. J. Am. chem. Soc. 110, 8651–8656 (1993).
Gerlt, J.A. & Gassman, P.G. An explanation for rapid enzyme-catalyzed proton abstraction from carbon acids: Importance of late transition states in concerted mechanisms. J. Am. chem. Soc. 115, 11552–11568 (1993).
Jenkins, J.A., Nasser, W., Scott, M., Pickersgill, R., Vignon, J.-C. & Robert-Baudouy, J. Crystallization and preliminary X-ray studies of the pectate lyase from Bacillus subtilis. J. molec. Biol. 228, 1255–1258 (1992).
Pickersgill, R.W., Harris, G.W. & Jenkins, J.A. Determination of the structure of Bacillus subtilis pectate lyase. Proceedings of the CCP4 Meeting “From First Map to Final Model” in the press (1994).
Howard, A.J. et al. The use of an image proportional counter in macromolecular crystallography. J. appl. Crystallogr. 20, 383–387 (1987).
CCP4 suite, Computer programmes for protein crystallography, (Daresbury Laboratory Warrington, England).
Jones, T.A., Zou, J-Y, Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these maps. Acta. crystallogr. A47, 110–119 (1991).
Zhang, K.Y.J. SQUASH-Combining constraints for macromolecular phase refinement and extension. Acta Crystallogr. D49, 213–222 (1993)
Brünger, A.T., Kuriyan, J. & Karplus, M. Crystallographic R factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Lamzin,V.S. & Wilson, K.S. Automated refinement of protein models. Acta. crystallogr. D49, 129–147 (1993).
Driessen, H. et al. Restrain: restrained structure factor least-squares refinement program for macromolecular structures. J. appl. crystallogr. 22, 510–516 (1989).
Kraulis, P.J. MOLSCRIPT; a program to produce both detailed and schematic plots of proteins. J. appl. Crystallogr. 24, 946–950 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Pickersgill, R., Jenkins, J., Harris, G. et al. The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat Struct Mol Biol 1, 717–723 (1994). https://doi.org/10.1038/nsb1094-717
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb1094-717
This article is cited by
-
Analysis of protein secretion in Bacillus subtilis by combining a secretion stress biosensor strain with an in vivo split GFP assay
Microbial Cell Factories (2023)
-
Pectinolytic lyases: a comprehensive review of sources, category, property, structure, and catalytic mechanism of pectate lyases and pectin lyases
Bioresources and Bioprocessing (2021)
-
Origins and features of pectate lyases and their applications in industry
Applied Microbiology and Biotechnology (2020)
-
Biochemical and Molecular Characterizations of a Novel pH- and Temperature-Stable Pectate Lyase from Bacillus amyloliquefaciens S6 for Industrial Application
Molecular Biotechnology (2019)
-
Structural insights into the molecular mechanisms of pectinolytic enzymes
Journal of Proteins and Proteomics (2019)