Abstract
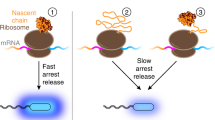
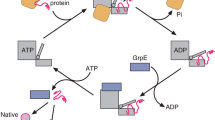
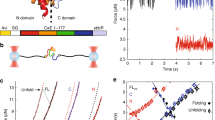
The 62 kDa protein firefly luciferase folds very rapidly upon translation on eukaryotic ribosomes. In contrast, the chaperone-mediated refolding of chemically denatured luciferase occurs with significantly slower kinetics. Here we investigate the structural basis for this difference in folding kinetics. We find that an N-terminal domain of luciferase (residues 1–190) folds co-translationally, followed by rapid formation of native protein upon release of the full-length polypeptide from the ribosome. In contrast sequential domain formation is not observed during in vitro refolding. Discrete unfolding steps, corresponding to domain unfolding, are however observed when the native protein is exposed to increasing concentrations of denaturant. Thus, the co-translational folding reaction bears more similarities to the unfolding reaction than to refolding from denaturant. We propose that co-translational domain formation avoids intramolecular misfolding and may be critical in the folding of multidomain proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Anfinsen, C.B. Principles that govern the folding of protein chains. Science 181, 223–30 (1973).
Creighton, T.E. Protein folding. (W.H. Freeman, New York; 1992).
Jaenicke, R. Protein folding: local structures, domains, subunits, and assemblies. Biochemistry 30, 3147–3160 (1991).
Dill, K.A. & Chan, H.S. From Levinthal to pathways to funnels Nature Struct. Biol. 4, 10– 19 (1997).
Dobson, C.M., Sali, A. & Karplus, M. Protein-folding: a perspective from theory and experiment. Angew. Chem. Int. Edn. Engl. 37, 868– 893 (1998).
Gething, M.-J. & Sambrook, J. Protein folding in the cell. Nature 355, 33–45 ( 1992).
Hartl, F.U. Molecular chaperones in cellular protein folding Nature 381, 571–579 (1996).
Ellis, R.J. The "bio" in biochemistry: protein folding inside and outside the cell. Science 272, 1448–1449 ( 1996).
Bukau, B. & Horwich, A.L. The Hsp70 and Hsp60 chaperone machines Cell 92, 351–366 (1998).
Netzer, W. & Hartl, F. Recombination of protein domains facilitated by co-translational folding in eukaryotes Nature 388 , 343–349 (1997).
Conti, E., Franks, N.P. & Brick, P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes Structure 4, 287–298 (1995).
McNew, J.A. & Goodman, J.M. The targeting and assembly of peroxisomal proteins: some old rules do not apply Trends Biochem. Sci. 21, 54–58 ( 1996).
deWet, J.R., Wood, K.V., DeLuca, M., Helinsky, D.R. & Subramani, S. Firefly luciferase gene: structure and expression in mammalian cells Mol. Cell. Biol. 7, 725– 737 (1987).
Nimmesgern, E. & Hartl, F.U. ATP-dependent protein refolding activity in reticulocyte lysate. Evidence for the participation of different chaperone components. FEBS Lett. 331, 25–30 (1993).
Schumacher, R.J. et al. ATP-Dependent chaperoning activity of reticulocyte lysate J. Biol. Chem. 269, 9493– 9499 (1994).
Freeman, B.C., Myers, M.P., Schumacher, R. & Morimoto, R.I. Identification of a regulatory motif In Hsp70 that affects ATPase activity; substrate-binding and interaction with Hdj-1 EMBO J. 14, 2281–2292 (1995).
Herbst, R., Schafer, U. & Seckler, R. Equilibrium intermediates in the reversible unfolding of firefly (Photinus pyralis) luciferase J. Biol. Chem. 272, 7099–7105 ( 1997).
Frydman, J., Nimmesgern, E., Ohtsuka, K. & Hartl, F.U. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones Nature 370, 111– 117 (1994).
Kolb, V.A., Makeyev, E.V. & Spirin, A.S. Folding of firefly luciferase during translation in a cell-free system EMBO J. 13, 3631– 3637 (1994).
Schneider, C. et al. Pharmacological shifting of a balance between protein refolding and degradation mediated by Hsp90 Proc. Natl. Acad. Sci. USA 93, 14536–14541 (1996).
Garel, J.R. In Protein folding (ed. Creighton, T.E.) 405–454 (W.H. Freeman, New York; 1992).
Taubes, G. Misfolding the way to disease Science 271, 1493–1495 (1996).
Mitraki, A., Fane, B., Haasepettingell, C., Sturtevant, J. & King, J. Global suppression of protein folding defects and inclusion body formation Science 253, 54–58 (1991).
Qu, B.H. & Thomas, P.J. Alteration of the cystic-fibrosis transmembrane conductance regulator folding pathway: effects of the delta-F508 mutation on the thermodynamic stability and folding yield of NBD1. J. Biol. Chem. 271, 7261–7264 (1996).
Lui, M., Tempst, P. & Erdjument-Bromage, H. Methodical analysis of protein–nitrocellulose interactions to design a refined digestion protocol. Anal. Biochem. 241, 156–166 ( 1996).
Elicone, C., Lui, M., Geromanos, S., Erdjument-Bromage, H. & Tempst, P. Microbore reversed-phase high-performance liquid chromatographic purification of peptides for combined chemical sequencing / laser-desorption mass spectrometric analysis. J. Chromatogr 676, 121–137 (1994).
Erdjument-Bromage, H., Lui, M., Sabatini, D.M., Snyder, S.H. & Tempst, P. High-sensitivity sequencing of large proteins: partial structure of the rapamycin-FKBP12 target. Protein Sci. 3, 2435–2446 (1994).
Tempst, P., et al. In Mass spectrometry in the biological sciences. (eds Burlingame, A.L. & Carr, S.A.) 105–133 (Humana Press, Totowa, New Jersey; 1996).
Acknowledgements
J.F. is supported by an NIH grant. P.T. is supported by an NSF grant and an NCI grant to the Sloan-Kettering Structural Chemistry Laboratory.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Frydman, J., Erdjument-Bromage, H., Tempst, P. et al. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat Struct Mol Biol 6, 697–705 (1999). https://doi.org/10.1038/10754
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/10754
This article is cited by
-
Universal protein misfolding intermediates can bypass the proteostasis network and remain soluble and less functional
Nature Communications (2022)
-
An epilepsy-causing mutation leads to co-translational misfolding of the Kv7.2 channel
BMC Biology (2021)
-
Interactions between nascent proteins and the ribosome surface inhibit co-translational folding
Nature Chemistry (2021)
-
Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein
Nature Communications (2020)
-
Non-equilibrium dynamics of a nascent polypeptide during translation suppress its misfolding
Nature Communications (2019)