Abstract
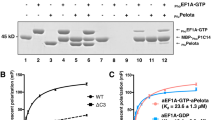
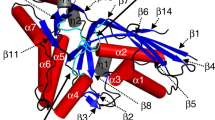
In the elongation cycle of protein biosynthesis, the nucleotide exchange factor eEF1Bα catalyzes the exchange of GDP bound to the G-protein, eEF1A, for GTP. To obtain more information about the recently solved eEF1A–eEF1Bα structure, we determined the structures of the eEF1A–eEF1Bα–GDP–Mg2+, eEF1A–eEF1Bα–GDP and eEF1A–eEF1Bα–GDPNP complexes at 3.0, 2.4 and 2.05 Å resolution, respectively. Minor changes, specifically around the nucleotide binding site, in eEF1A and eEF1Bα are consistent with in vivo data. The base, sugar and α-phosphate bind as in other known nucleotide G-protein complexes, whereas the β- and γ-phosphates are disordered. A mutation of Lys 205 in eEF1Bα that inserts into the Mg2+ binding site of eEF1A is lethal. This together with the structures emphasizes the essential role of Mg2+ in nucleotide exchange in the eEF1A–eEF1Bα complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Merrick, W.C. & Nyborg, J. In Translational control of gene expression (eds Sonenberg, N., Hershey, J.W.B. & Mathews, M.B.), 89–126 (Cold Spring Harbor Laboratory Press, New York; 2000).
Edmonds, B.T., Bell, A., Wyckoff, J., Condeelis, J. & Leyh, T.S. J. Biol. Chem. 273, 10288–10295 (1998).
Janssen, G.M. & Moller, W. J. Biol. Chem. 263, 1773–1778 (1988).
Andersen G. et al. Mol. Cell 6, 1261–1266 (2000).
Kawashima, T., Bethet-Colominas, C., Wulff, M., Cusack, S. & Leberman, R. Nature 379, 511–518 (1996).
Wang, Y., Jiang, Y., Meyering-Voss, M., Sprinzl, M. & Sigler, P.B. Nature Struct. Biol. 4, 650–656 (1997).
Goldberg, J. Cell 95, 237–248 (1998).
Boriack-Sjodin, P.A., Margarit, S.M., Bar-Sagi, D. & Kuriyan, J. Nature 394, 337–343 (1998).
Worthylake D.K., Rossman, K.L. & Sondek, J. Nature 408, 682–688 (2000).
Sprang, S.R. & Coleman, D.E. Cell 95, 155–158 (1998).
Carr-Schmid, A., Durko, N., Cavallius, J., Merrick, W.C. & Kinzy, T.G. J. Biol. Chem. 274, 30297–30302 (1999).
Carr-Schmid, A. et al. Mol. Cell Biol. 19, 5257–5266 (1999).
Dinman, J.D. & Kinzy, T.G. RNA 3, 870–881 (1997).
Sandbaken, M.G. & Culbertson, M.R. Genetics 141, 481–489 (1995).
Kinzy, T.G., Woolford, J.L., Jr. Genetics 141, 481–489 (1995).
Pan, J.Y. & Wessling-Resnick, M. Bioessays 20, 516–521 (1998).
John, J. et al. J. Biol. Chem. 268, 923–929 (1993).
Cherfils, J. & Chardin, P. Trends Biochem. Sci. 24, 306–311 (1999).
Wittinghofer, F. Nature 394, 317–320 (1998).
Beraud-Dufour, S. et al. EMBO J. 17, 3651–3659 (1998).
Van Damme, H.T., Amons, R. & Moller, W. Eur. J. Biochem. 207, 1025–1034 (1992).
Pedersen, L.P., Andersen, G.R., Knudsen, C.K., Kinzy, T.G. & Nyborg, J. Acta Crystallogr. D 57, 159–161 (2000).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1997).
Brunger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Cowan, S., Zou, J.-Y. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 (1991).
Koradi, R., Billeter, M. & Wuthrich, K. J. Mol. Graph. 14, 29–32 (1996).
Boeke, J.D., Trueheart, J., Natsoulis, G. & Fink, G.R. Methods Enzymol. 154, 164–175 (1987).
Sprang, S.R. Annu. Rev. Biochem. 66, 629–678 (1997).
Acknowledgements
We are grateful to K. Djinovic at ELETTRA, Trieste, for help during data collection. GRA was supported by Aarhus University. J.N. was supported by the Program for Biotechnological Research of the Danish Natural Science Research Council, DANSYNC, and the EU. T.G.K. was supported by the NIH and the NSF and L.V. by NIH.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Andersen, G., Valente, L., Pedersen, L. et al. Crystal structures of nucleotide exchange intermediates in the eEF1A–eEF1Bα complex. Nat Struct Mol Biol 8, 531–534 (2001). https://doi.org/10.1038/88598
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/88598
This article is cited by
-
The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates
Nature Communications (2018)
-
Elongation factor Tu is a multifunctional and processed moonlighting protein
Scientific Reports (2017)
-
Structural rationale for the cross-resistance of tumor cells bearing the A399V variant of elongation factor eEF1A1 to the structurally unrelated didemnin B, ternatin, nannocystin A and ansatrienin B
Journal of Computer-Aided Molecular Design (2017)
-
The C-terminal region of human eukaryotic elongation factor 1Bδ
Journal of Biomolecular NMR (2016)
-
Altered machinery of protein synthesis is region- and stage-dependent and is associated with α-synuclein oligomers in Parkinson’s disease
Acta Neuropathologica Communications (2015)