Abstract
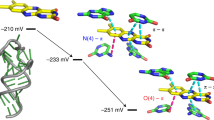
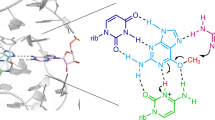
The leadzyme is a small RNA motif that catalyzes a site-specific, Pb2+-dependent cleavage reaction. As such, it is an example of a metal-dependent RNA enzyme. Here we describe the X-ray crystallographic structure of the leadzyme, which reveals two independent molecules per asymmetric unit. Both molecules feature an internal loop in which a bulged purine base stack twists away from the helical stem. This kinks the backbone, rendering the phosphodiester bond susceptible to cleavage. The independent molecules have different conformations: one leadzyme copy coordinates Mg2+, whereas the other binds only Ba2+ or Pb2+. In the active site of the latter molecule, a single Ba2+ ion coordinates the 2´-OH nucleophile, and appears to mimic the binding of catalytic lead. These observations allow a bond cleavage reaction to be modeled, which reveals the minimal structural features necessary for catalysis by this small ribozyme.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Gilbert, W. The RNA world. Nature 319, 618 (1986).
Cech, T.R. in The RNA world (eds. Gesteland, R.F. & Atkins, J.F.) 239– 269 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York; 1993).
Noller, H.F., Hoffarth, V. & Zimniak, L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256, 1416 –1419 (1992).
Nitta, I., Kamada, Y., Noda, H., Ueda, T. & Watanabe, K. Reconstitution of peptide bond formation with Escherichia coli 23S ribosomal RNA domains. Science 281, 666–669 (1998).
Long, D.M. & Uhlenbeck, O.C. Self-cleaving catalytic RNA. FASEB J. 7, 25–30 (1993).
Pley, H.W., Flaherty, K.M. & McKay, D.B. Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74 (1994).
Scott, W.G., Finch, J.T. & Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell 81, 991–1002 (1995).
Scott, W.G., Murray, J.B., Arnold, J., Stoddard, B.L. & Klug, A. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science 274, 2065 –2069 (1996).
Murray, J.B. et al. The structural basis of hammerhead ribozyme self-cleavage. Cell 92, 665–673 (1998).
Wedekind, J.E. & McKay, D.B. Crystallographic structures of the hammerhead ribozyme: relationship to ribozyme folding and catalysis. Ann. Rev. Biophys. Biomol. Struct. 27, 475–502 (1998).
Dirheimer, G. et al. Primary structure of transfer RNA. Biochimie 54, 127–144 (1972).
Brown, R.S., Hingerty, B.E., Dewan, J.C. & Klug, A. Pb(II)-catalysed cleavage of the sugar–phosphate backbone of yeast tRNAPhe—implications for lead toxicity and self-splicing RNA. Nature 303 , 543–546 (1983).
Pan, T. & Uhlenbeck, O.C. A small metalloribozyme with a two-step mechanism. Nature 358, 560– 563 (1992).
Hodel, A., Kim, S.-H. & Brünger, A.T. Model bias in macromolecular crystal structures. Acta Crystallogr. A 48, 851–859 (1992).
Adams, P.D., Pannu, N.S., Read, R.J. & Brünger, A.T. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. USA 94, 5018– 5023 (1997).
Sugimoto, N. & Ohmichi, T. Site-specific cleavage reaction catalyzed by leadzyme is enhanced by combined effect of lead and rare earth ions. FEBS Lett. 393, 97– 100 (1996).
Brown, R.S., Dewan, J.C. & Klug, A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry 24, 4785–4801 ( 1985).
Gao, Y.G., Robinson, H., van Boom, J.H. & Wang, A.H. Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA. Biophys. J. 69, 559– 568 (1995).
Gao, Y.G., Sriram, M. & Wang, A.H. Crystallographic studies of metal ion–DNA interactions: different binding modes of cobalt(II), copper(II) and barium(II) to N7 of guanines in Z-DNA and a drug–DNA complex. Nucleic Acids Res. 21, 4093–4101 ( 1993).
Hoogstraten, C.G., Legault, P. & Pardi, A. NMR solution structure of the lead-dependent ribozyme: evidence for dynamics in RNA catalysis. J. Mol. Biol. 284, 337–350 (1998).
Legault, P., Hoogstraten, C.G., Metlitzky, E. & Pardi, A. Order, dynamics and metal binding in the lead-dependent ribozyme. J. Mol. Biol. 284, 325–335 (1998).
Pan, T., Dichtl, B. & Uhlenbeck, O.C. Properties of an in vitro selected Pb2+ cleavage motif. Biochemistry 33, 9561–9565 (1994).
Chartrand, P., Usman, N. & Cedergren, R. Effect of structural modifications on the activity of the leadzyme. Biochemistry 36, 3145– 3150 (1997).
Legault, P. & Pardi, A. Unusual dynamics and pKa shift at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc. 119, 6621–6628 ( 1995).
Saenger, W. Principles of nucleic acid structure (Springer-Verlag, New York; 1984).
Lemieux, S., Chartrand, P., Cedergren, R. & Major, F. Modeling active RNA structures using the intersection of conformational space: application to the lead-activated ribozyme. RNA 4, 739–749 (1998).
Wedekind, J.E. & McKay, D.B. Purification, crystallization, and X-ray diffraction analysis of small ribozymes. Methods Enzymol. in the press (1999).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 ( 1997).
Terwilliger, T.C. SOLVE: an automated crystallographic structure solution program for MIR and MAD. Edition 1.04 (www.solve.lanl.gov, Los Alamos National Laboratory; 1997).
de La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength diffraction methods. Methods Enzymol. 276, 472–494 (1997).
Abrahams, J.P. & Leslie, A.G.W. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 42, 30–42 (1996).
Jones, T.A., Zhou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved Methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Pannu, N.S. & Read, R.J. Improved structure refinement through maximum likelihood. Acta Crystallogr. A 52, 659–668 (1996).
Brünger, A.T., Kuriyan, J.M. & Karplus, M. Crystallographic R factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Jiang, J.S. & Brünger, A.T. Protein hydration observed by X-ray diffraction. Solvation properties of penicillopepsin and neuraminidase crystal structures. J. Mol. Biol. 243, 100 –115 (1994).
Sheriff, S. & Hendrickson, W.A. Description of overall anisotropy in diffraction from macromolecular crystals. Acta Crystallogr. A 43, 118–121 ( 1987).
Read, R.J. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42, 140 –149 (1986).
Esnouf, R.M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15, 133–138 (1997).
Merritt, E.A. & Bacon, D.J. Raster 3D: photorealistic molecular graphics. Methods Enzymol. 277, 505– 524 (1997).
Acknowledgements
We thank C. Kielkopf, J. Puglisi, W. Weis, M. Sousa, and A. Kolatkar for helpful discussions, as well as expert technical advice and support; C. Trinh and the staff of SSRL for assistance with the X-ray data collection; also, we thank A. Pardi and C. Hoogstraten for discussions and sharing their unpublished results on the solution structure of the leadzyme. J.E.W. is a Burroughs Wellcome Fund Fellow of the Life Sciences Research Foundation (LSRF). This work was supported by funds from the LSRF and an NIH grant to D.B.M. Operation of SSRL beamlines 7-1 and 9-1 is supported by the DOE, Office of Basic Energy Sciences. The Biotechnology Program at SSRL is supported by the NIH, National Center for Research Resources Biomedical Technology Program and the DOE, Office of Biological and Environmental Research.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wedekind, J., McKay, D. Crystal structure of a lead-dependent ribozyme revealing metal binding sites relevant to catalysis. Nat Struct Mol Biol 6, 261–268 (1999). https://doi.org/10.1038/6700
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/6700
This article is cited by
-
Concerted dynamics of metallo-base pairs in an A/B-form helical transition
Nature Communications (2019)
-
Crystal structure of an RNA-cleaving DNAzyme
Nature Communications (2017)
-
Searching for a DNAzyme Version of the Leadzyme
Journal of Molecular Evolution (2015)
-
Theoretical analysis of noncanonical base pairing interactions in RNA molecules
Journal of Biosciences (2007)
-
Cleavage of RNA bulge loops by artificial RNases
Russian Chemical Bulletin (2006)