Abstract
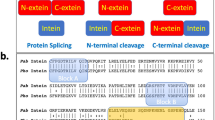
Several genes from prokaryotes and lower eukaryotes have been found to contain an in-frame open reading frame, which encodes for an internal protein (intein). Post-translationally, the internal polypeptide auto-splices and ligates the external sequences to yield a functional external protein (extein) and an intein. Most, but not all inteins, contain, apart from a splicing domain, a separate endonucleolytic domain that enables them to maintain their presence by a homing mechanism. We report here the crystal structure of an intein found in the gyrase A subunit from Mycobacterium xenopi at 2.2 Å resolution. The structure contains an unusual β-fold with the catalytic splice junctions at the ends of two adjacent antiparallel β-strands. The arrangement of the active site residues Ser 1, Thr 72, His 75, His 197, and Asn 198 is consistent with a four-step mechanism for the cleavage–ligation reaction. Using site-directed mutagenesis, the N-terminal cysteine, proposed as the nucleophile in the first step of the splicing reaction, was changed to a Ser 1 and Ala 0, thus capturing the intein in a pre-spliced state.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Perler, F.B. et al. Protein splicing elements: Inteins and exteins—a definition of terms and recommended nomenclature. Nucleic Acids Res. 22, 1125–1127 (1994).
Cooper, A.A. & Stevens, T.H. Protein splicing: self-splicing of genetically mobile elements at the protein level. Trends Biochem. Sci. 20, 351–356 (1995).
Guan, C. et al. Activation of glycosylasparaginase. Formation of active N-terminal threonine by intramolecular autoproteolysis. J. Biol. Chem. 271, 1732–1737 (1996).
Porter, J.A. et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86, 21–34 (1996).
van Poelje, P.D. & Snell, E.E. Pyruvoyl-dependent enzymes. Annu. Rev. Biochem. 59, 29–59 (1990).
Kane, P.M. et al. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H+-adenosine triphosphatase. Science 250, 651–657 (1990).
Perler, F.B., Olsen, G.J. & Adams, E. Compilation and analysis of intein sequences. Nucleic Acids Res. 25, 1087–1094 (1997).
Gimble, F.S. & Thorner, J. Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature 357, 301–306 (1992).
Bremer, M.C.D., Gimble, F.S., Thorner, J. & Smith, C.L. VDE endonuclease cleaves Saccharomyces cerevisiae genomic DNA at a single site: physical mapping of the VMA1 gene. Nucleic Acids Res. 20, 5484 (1992).
Pietrokovski, S. Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Sci. 3, 2340–2350 (1994).
Perler, F.B. et al. Intervening sequences in an archaea DNA polymerase gene. Proc. Natl. Acad. Sci. USA 89, 5577–5581 (1992).
Duan, X., Gimble, F.S. & Quiocho, F.A. Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell 89, 555–564 (1996).
Xu, M.-Q. & Perler, F.B. The mechanism of protein splicing and its modulation by mutation. EMBO J. 15, 5146–5153 (1996).
Davis, E.O. et al. Protein splicing in the maturation of M. tuberculosis recA protein: a mechanism for tolerating a novel class of intervening sequence. Cell 71, 201–210 (1992).
Hirata, R. & Anraku, Y. Mutations at the putative junction sites of the yeast VMA1 protein, the catalytic subunit of the vacuolar membrane H+-ATPase, inhibit its processing by protein splicing. Biochem. Biophys. Res. Comm. 188, 40–47 (1992).
Cooper, A.A., Chen, Y., Lindorfer, M.A. & Stevens, T.H. Protein splicing of the yeast TFP1 intervening protein sequence: a model of self-excision. EMBO J. 12, 2575–2583 (1993).
Xu, M.-Q. et al. Protein splicing: an analysis of the branched intermediate and its resolution by succinimide formation. EMBO J. 13, 5517–5522 (1994).
Dawson, P.E., Muir, T.W., Clark-Lewis, I. & Kent, S.B. Synthesis of proteins by native chemical ligation. Science 266, 776–779 (1994).
Storer, A.C. & Ménard, R. Catalytic mechanism in papain family of cysteine peptidases. Meth. Enz. 244, 486–500 (1994).
Drenth, J., Jansonius, J.N., Koekoek, R., Swen, H.M. & Wolthers, B.G. Structure of papain. Nature 218, 929–932 (1968).
Ramachandran, G.N. & Mitra, A.K. An explanation for the rare occurrence of cis peptide units in proteins and polypeptides. J. Mol. Biol. 107, 85–92 (1976).
Fsihi, H., Vincent, V. & Cole, S.T. Homing events in the gyrA gene of some mycobacteria. Proc. Natl. Acad. Sci. USA 93, 3410–3415 (1996).
Oinonen, C., Tikkanen, R., Rouvinen, J. & Peltonen, L. Three-dimensional structure of human lyosomal aspartylglucosaminidase. Nature Struct. Biol. 2, 1102–1107 (1995).
Smith, J.L. et al. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science 264, 1427–1433 (1994).
Duggleby, H.J. et al. Penicillin acylase has a single-amino-acid catalytic center. Nature 373, 264–268 (1995).
Löwe, J. et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science 268, 533–539 (1995).
Brannigan, J.A. et al. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378, 416–419 (1995).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data in oscillation mode. Meth. Enz. 276, 307–326 (1997).
Collaborative Computational Project No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Furey, W. & Swaminathan, S. PHASES. Am. Crystallogr. Assoc. Annu. Mtg. Program. Abstr. 18, 73 (1990).
Jones, T.A., Zou, J.-Y., Kjelgaard, M. & Cowan, S.W. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A. X-PLOR Version 3.1: A System for X-ray crystallography and NMR. (Yale University Press, New Haven, Connecticut; 1992).
Evans, S.V. SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics 11, 134–138 (1993).
Hall, T.M.T. et al. Crystal structure of a hedgehog autoprocessing domain: homology between hedgehog and self-splicing proteins. Cell 91, 85–97 (1997).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Klabunde, T., Sharma, S., Telenti, A. et al. Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat Struct Mol Biol 5, 31–36 (1998). https://doi.org/10.1038/nsb0198-31
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0198-31
This article is cited by
-
An alternative domain-swapped structure of the Pyrococcus horikoshii PolII mini-intein
Scientific Reports (2021)
-
Structural basis for RING-Cys-Relay E3 ligase activity and its role in axon integrity
Nature Chemical Biology (2020)
-
Exploring chemoselective S-to-N acyl transfer reactions in synthesis and chemical biology
Nature Communications (2017)
-
Comparative Analysis of the Effectiveness of C-terminal Cleavage Intein-Based Constructs in Producing a Recombinant Analog of Anophelin, an Anticoagulant from Anopheles albimanus
Applied Biochemistry and Biotechnology (2015)
-
Backbone assignments of mini-RecA intein with short native exteins and an active N-terminal catalytic cysteine
Biomolecular NMR Assignments (2015)