Abstract
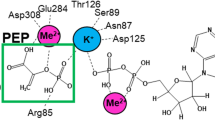
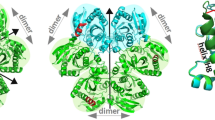
The crystal structure of the phosphoglycerate dehydrogenase from Eschehchia coli is unique among dehydrogenases. It consists of three clearly separate domains connected by flexible hinges. The tetramer has approximate 222 symmetry with the principal contacts between the subunits forming between either the nucleotide binding domains or the regulatory domains. Two slightly different subunit conformations are present which vary only in the orientations of the domains. There is a hinge-like arrangement near the active site cleft and the serine effector site is provided by the regulatory domain of each of two subunits. Interdomain flexibility may play a key role in both catalysis and allosteric inhibition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Monod, J., Wyman, J. & Changeux, J.-P. On the nature of allosteric transitions: A plausible model, J. molec. Biol. 12, 88–118 (1965).
Ichihara, A. & Greenberg, D.M. Further studies on the pathway of serine formation from carbohydrate, J. biol. Chem. 224, 331–340 (1957).
Pizer, L.I. The pathway and control of serine biosynthesis in Escherichia coli, J. biol. Chem. 238, 3934–3944 (1963).
Willis, J.E. & Sallach, H.J. The occurence of D-3-phosphoglycerate dehydrogenase in animal tissues, Biochim. biophys. Acta 81, 39–54 (1964).
Slaughter, J.C. & Davies, D.D. Inhibition of 3-phosphoglycerate dehydrogenase by L-serine, Biochem J. 109, 749–755 (1968).
Tobey, K.L. & Grant, G.A. The nucleotide sequence of the serA gene of Escherichia coli and the amino acid sequence of the encoded protein, D-3-phosphoglycerate dehydrogenase, J. biol. Chem. 261, 12179–12183 (1986).
Grant, G.A. A new family of 2-hydroxyacid dehydrogenases, Biochem. biophys. Res. Comm. 165, 1371–1374 (1989).
Goldberg, J.D., Yoshida, T. & Brick, P. Crystal structure of a NAD-dependent D-Glycerate dehydrogenase at 2.4 Å resolution, J. molec. Biol. 236, 1123–1140 (1994).
Lamzin, V.S., Dauter, Z., Popov, V.O., Harutyunyan, E.H. & Wilson, K.S. High resolution structures of holo and apo formate dehydrogenase, J. molec. Biol. 236, 759–785 (1994).
Sugimoto, E. & Pizer, L.I. The mechanism of end product inhibition of serine biosynthesis: I. purification and kinetics of phosphoglycerate dehydrogenase, J. biol. Chem. 243, 2081–2089 (1968).
Sugimoto, E. & Pizer, L.I. The mechanism of end product inhibition of serine biosynthesis: II. optical studies of phosphoglycerate dehydrogenase, J. biol. Chem. 243, 2090–2098 (1968).
Dubrow, R. & Pizer, L.I. Transient kinetic studies on the allosteric transition of phosphoglycerate dehydrogenase, J. biol. Chem. 252, 1527–1538 (1977).
Dubrow, R. & Pizer, L.I. Transient kinetic and deuterium isotope effect studies on the catalytic mechanism of phosphoglycerate dehydrogenase, J. biol. Chem. 252, 1539–1551 (1977).
Birktoft, J.J. & Banaszak, L.J. The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases, J. biol. Chem. 258, 472–482 (1983).
Janin, J. Shared structural motif in proteins Nature 365, 21 1993.
Dumas, C et al., X-ray structure of nucleoside diphosphate kinase, EMBO J. 11, 3203–3208 (1992).
Kosman, R.P., Gouaux, J.E. & Lipscomb, W.N. Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 Å resolution: implications for ATCase mutants and the mechanism of negative cooperativity, Proteins 15, 147–176 (1993).
Schuller, D.J., Fetter, C.H., Banaszak, L.J. & Grant, G.A. Enhanced expression of the Escherichia coli serA gene in a plasmid vector: purification, crystallization, and preliminary X-ray data of D-3-phosphoglycerate dehydrogenase, J. biol. Chem. 264, 2645–2648 (1989).
Howard, A.J., Gilliland, G.L., Finzel, B.C., Poulos, T.L., Ohlendorf, D.H. & Salemme, F.R. The use of an imaging proportional counter in macromolecular crystallography J. Appl. Crystallogr. 20, 383–387 (1987).
Terwilliger, T.C. & Eisenberg, D. Unbiased three-dimensional refinement of heavy-atom parameters by correlation of origin-removed Patterson functions, Acta crystallogr. A39, 813–817 (1983).
Terwilliger, T.C. Kim, S.-H. Eisenberg, D. Generalized method of determining heavy-atom positions using the difference Patterson function, Acta Crystallogr. A43, 1–5 (1987).
Cowtan, K.D. & Main, P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints, Acta crystallogr. D49, 148–157 (1993).
Levitt, D.G. & Banaszak, L.J. A new routine for thinning, editing and fitting MIR maps using real space molecular dynamics, J. appl. Crystallogr. 26, 736–745 (1993).
Brunger, A.T., Kuriyan, J. & Karplus, M. R-factor refinement by molecular dynamics, Science 235, 458–460 (1987).
Bio-Graphics, Turbo-Frodo (Marseille 1992).
Jones, T.A. Interactive computer graphics: FRODO in Meths Enzymol. 115, 157–171 (1985).
Collaborative Computational Project, Number 4, The CCP4 suite: programs for protein crystallography, Acta crystallogr D50, 760–763 (1994)
Lee, B. & Richards, F.M. The interpretation of protein structures: Estimation of static accessibility, J. molec. Biol. 55, 379–400 (1971)
Read, R. Improved fourier coefficients for maps using phases from partial structures with errors, Acta crystallogr. A42, 140–149 (1986)
Bernstein, F.C. et al. The protein data bank: a computer-based archival file for macromolecular structures. J. molec. Biol. 112, 535–542 (1977).
Kraulis, P.J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures, J. appl. Crystallogr. 24, 946–950 (1991).
Evans, S.V. SETOR: Hardware-lighted three-dimensional solid model representations of macromolecules J. molec. Graphics 11, 134–138 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Schuller, D., Grant, G. & Banaszak, L. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat Struct Mol Biol 2, 69–76 (1995). https://doi.org/10.1038/nsb0195-69
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0195-69
This article is cited by
-
A morpheein equilibrium regulates catalysis in phosphoserine phosphatase SerB2 from Mycobacterium tuberculosis
Communications Biology (2023)
-
Cloning, heterologous expression, and functional characterization of a hydroxyphenylpyruvate reductase (HPPR) gene involved in rosmarinic acid biosynthesis in Prunella vulgaris
Plant Cell, Tissue and Organ Culture (PCTOC) (2023)
-
3-Phosphoglycerate dehydrogenase: a potential target for cancer treatment
Cellular Oncology (2021)
-
The lineage and diversity of putative amino acid sensor ACR proteins in plants
Amino Acids (2020)
-
Classification, substrate specificity and structural features of D-2-hydroxyacid dehydrogenases: 2HADH knowledgebase
BMC Evolutionary Biology (2018)