Abstract
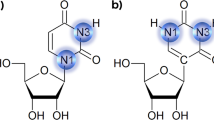
The crystal structure of the RNA dodecamer 5′-GGACUUUGGUCC-3′ has been determined from X-ray diffraction data to 2.6 Å resolution. This oligomer forms an asymmetric double helix in the crystal. Four consecutive non-Watson-Crick base-pairs are formed in the middle of the duplex including the first intrahelical U-U (or T-T) pairs observed in an oligonucleotide crystal structure. Two different conformations of U-U pairs are observed in the context of the surrounding sequence. One of these pairs is highly twisted, allowing a bound water to bridge across strands in the major groove. The crystal packing illustrates a new form of RNA helix-helix interaction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Young, L.S. et al. A class III transcription factor composed of RNA. Science 252, 542–546 (1991).
Hingerty, B., Brown, R.S. & Jack, A. Further refinement of the structure of yeast tRNAphe J. molec. Biol. 124, 523–534 (1978).
Sussman, J.L., Holbrook, S.R., Warrant, R.W., Church, G.M. & Kim, S -H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J. molec. Biol. 123, 607–630 (1978).
Comarmond, M.B., Giege, R., Thierry, J.C., Moras, D. & Fischer, J. Three-dimensional structure of yeast tRNA(asp). I. Structure determination. Acta crystallogr. B42, 272–280 (1986).
Hou, Y.-M. & Schimmel, P. A simple structural feature is a major determinant of the identity of a transfer RNA Nature 333, 140–145 (1988).
Olsen, H.S., Nelbrock, P., Cochrane, A.W. & Rosen, C.A. Secondary structure is the major determinant for interaction of HIV rev protein with RNA Science 247, 845–848 (1990).
Holbrook, S.R., Cheong, C., Tinoco, I., Jr & Kim, S. -H. . Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs Nature 353, 579–581 (1991).
Dock-Bregeon, A.C. et al. Crystallographic structure of an RNA helix: [U(UA)6A]2 J. molec. Biol. 209, 459–474 (1989).
Cruse, W.B.T. et al. Structure of a mispaired RNA double helix at 1.6 Å resolution and implications for the prediction of RNA secondary structure. Proc. natn. Acad. Sci. U.S.A. 91,4160–4164 (1994).
Kouchakdjian, M., Li, B.F.L., Swann, P.F. & Patel, D.J. Pyrimidine-pyrimidine base-pair mismatches in DNA: A nuclear magnetic resonance study of T-T pairing at neutral pH and C-C pairing at acidic pH in dodecanucleotide duplexes. J.molec. Biol. 202, 139–155 (1988).
Santa Lucia, J., Jr, Kierzek, R. & Turner, D.H. Stabilities of consecutive A-C, C-C, G-G, U-C, and U-U mismatches in RNA internal loops: Evidence for stable hydrogen-bonded U-U and C-C+ pairs. Biochemistry. 30, 8243–8250 (1991).
Venable, R.M., Widmalm, G., Brooks, B.R., Egan, W. & Pastor, R.W. Conformational states of a TT mismatch from molecular dynamics simulation of duplex d(CGCGATTCGCG) Biopolymers 32, 783–794 (1992).
Hunter, W.N., et al. The structure of guanosine-thymidine mismatches in B-DNA at 2.5 Å resolution. J. biol. Chem. 262, 9962–9970 (1987).
Brown, T.C. & Jiricny, J. Repair of base-base mismatches in simian and human cells. Genome 31, 578–583 (1989).
Fang, W. & Modrich, P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J. biol. Chem. 268, 11838–11844 (1993).
Wimberly, B., Varani, G. & Tinoco, I.J. The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 32, 1078–1087 (1993).
Brunger, A.T. Extension of molecular replacement: A new search strategy based on Patterson correlation refinement. Acta crystallogr. A46, 46–57 (1990).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Baeyens, K., De Bondt, H. & Holbrook, S. Structure of an RNA double helix including uracil-uracil base pairs in an internal loop. Nat Struct Mol Biol 2, 56–62 (1995). https://doi.org/10.1038/nsb0195-56
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0195-56
This article is cited by
-
Divalent cations can control a switch-like behavior in heterotypic and homotypic RNA coacervates
Scientific Reports (2019)
-
The generic geometry of helices and their close-packed structures
Theoretical Chemistry Accounts (2010)
-
Non-Watson Crick base pairs might stabilize RNA structural motifs in ribozymes — a comparative study of group-I intron structures
Journal of Biosciences (2003)
-
Solution structure of the tobramycin–RNA aptamer complex
Nature Structural Biology (1998)
-
The structure of r(UUCGCG) has a 5′-UU-overhang exhibiting Hoogsteen-like trans U•U base pairs
Nature Structural Biology (1996)