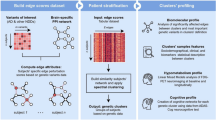
Key Points
-
Genetic findings in late-onset Alzheimer disease (AD) have not yet resulted in strategies to prevent or treat AD
-
Examining the position of genes carrying disease-associated variants in large-scale molecular networks can aid identification of coherent disease mechanisms
-
Not all network approaches are equal: recent approaches involve networks that are directed (with causal links), and specific to the tissue and disease state
-
AD neuropathology and AD-associated genetic variants decrease efficiency of information transfer in the brain connectome, which can be quantified by measuring structural and functional patterns in brain networks
-
Construction of multiscale models of the effects of AD-associated genetic variants with large neuronal simulations is now feasible, and will be useful to understand the effects of such variants and to screen therapeutics in silico
Abstract
Genetic studies in late-onset Alzheimer disease (LOAD) are aimed at identifying core disease mechanisms and providing potential biomarkers and drug candidates to improve clinical care of AD. However, owing to the complexity of LOAD, including pathological heterogeneity and disease polygenicity, extraction of actionable guidance from LOAD genetics has been challenging. Past attempts to summarize the effects of LOAD-associated genetic variants have used pathway analysis and collections of small-scale experiments to hypothesize functional convergence across several variants. In this Review, we discuss how the study of molecular, cellular and brain networks provides additional information on the effects of LOAD-associated genetic variants. We then discuss emerging combinations of these omic data sets into multiscale models, which provide a more comprehensive representation of the effects of LOAD-associated genetic variants at multiple biophysical scales. Furthermore, we highlight the clinical potential of mechanistically coupling genetic variants and disease phenotypes with multiscale brain models.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ferri, C. P. et al. Global prevalence of dementia: a Delphi consensus study. Lancet 366, 2112–2117 (2006).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Jack, C. R. et al. Introduction to the recommendations from the National Institute on Aging–Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 7, 257–262 (2011).
Dunkin, J. J. & Anderson-Hanley, C. Dementia caregiver burde: a review of the literature and guidelines for assessment and intervention. Neurology 51, S53–S60 (1998).
Hurd, M. D., Martorell, P., Delavande, A., Mullen, K. J. & Langa, K. M. Monetary costs of dementia in the United States. N. Engl. J. Med. 368, 1326–1334 (2013).
Langa, K. M. et al. National estimates of the quantity and cost of informal caregiving for the elderly with dementia. J. Gen. Intern. Med. 16, 770–778 (2001).
Hebert, L. E., Weuve, J., Scherr, P. A. & Evans, D. A. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology 80, 1778–1783 (2013).
Gatz, M. et al. Heritability for Alzheimer's disease: the study of dementia in Swedish twins. J. Gerontol. A Biol. Sci. Med. Sci. 52, M117–M125 (1997).
Janssen, J. et al. Early onset familial Alzheimer's disease mutation frequency in 31 families. Neurology 60, 235–239 (2003).
Campion, D. et al. Mutations of the presenilin I gene in families with early-onset Alzheimer's disease. Hum. Mol. Genet. 4, 2373–2377 (1995).
Barnes, D. E. & Yaffe, K. The projected effect of risk factor reduction on Alzheimer's disease prevalence. Lancet Neurol. 10, 819–828 (2011).
Lambert, J.-C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458 (2013).
Karch, C. M., Cruchaga, C. & Goate, A. M. Alzheimer's disease genetics: from the bench to the clinic. Neuron 83, 11–26 (2014).
Bertram, L. & Tanzi, R. E. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat. Rev. Neurosci. 9, 768–778 (2008).
Schellenberg, G. D. & Montine, T. J. The genetics and neuropathology of Alzheimer's disease. Acta Neuropathol. 124, 305–323 (2012).
Sleegers, K. et al. The pursuit of susceptibility genes for Alzheimer's disease: progress and prospects. Trends Genet. 26, 84–93 (2010).
Gillis, J. & Pavlidis, P. The impact of multifunctional genes on 'guilt by association' analysis. PLoS ONE 6, e17258 (2011).
Gillis, J. & Pavlidis, P. Assessing identity, redundancy and confounds in Gene Ontology annotations over time. Bioinformatics 29, 476–482 (2013).
Khatri, P., Sirota, M. & Butte, A. J. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 8, e1002375 (2012).
Parikshak, N. N., Gandal, M. J. & Geschwind, D. H. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 16, 441–458 (2015).
Gaiteri, C., Ding, Y., French, B., Tseng, G. C. & Sibille, E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 13, 13–24 (2014). This article summarizes the origin of coexpression networks — one of the most common types of molecular network studied in the context of AD.
Karch, C. M. & Goate, A. M. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 77, 43–51 (2015).
Goate, A. et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 349, 704–706 (1991).
Levy-Lahad, E. et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science 269, 973–977 (1995).
Rogaev, E. et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature 376, 775–778 (1995).
Corder, E. et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261, 921–923 (1993).
Cummings, J. L., Morstorf, T. & Zhong, K. Alzheimer's disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res. Ther. 6, 37 (2014).
Reiman, E. M. et al. Alzheimer's Prevention Initiative: a plan to accelerate the evaluation of presymptomatic treatments. J. Alzheimers Dis. 26, 321 (2011).
Bateman, R. J. et al. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N. Engl. J. Med. 367, 795–804 (2012).
Roses, A. D. et al. New applications of disease genetics and pharmacogenetics to drug development. Curr. Opin. Pharmacol. 14, 81–89 (2014).
Reisberg, B. et al. A 24-week open-label extension study of memantine in moderate to severe Alzheimer disease. Arch. Neurol. 63, 49–54 (2006).
Schneider, L. S., Dagerman, K. S., Higgins, J. P. & McShane, R. Lack of evidence for the efficacy of memantine in mild Alzheimer disease. Arch. Neurol. 68, 991–998 (2011).
Doody, R. S. et al. Phase 3 trials of solanezumab for mild-to-moderate Alzheimer's disease. N. Engl. J. Med. 370, 311–321 (2014).
Salloway, S. et al. Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer's disease. N. Engl. J. Med. 370, 322–333 (2014).
Holmes, C. et al. Long-term effects of Aβ42 immunisation in Alzheimer's disease: follow-up of a randomised, placebo-controlled phase I trial. Lancet 372, 216–223 (2008).
Rinne, J. O. et al. 11C-PiB PET assessment of change in fibrillar amyloid-β load in patients with Alzheimer's disease treated with bapineuzumab: a phase 2, double-blind, placebo-controlled, ascending-dose study. Lancet Neurol. 9, 363–372 (2010).
Jack, C. R. Jr. et al. Age-specific population frequencies of cerebral β-amyloidosis and neurodegeneration among people with normal cognitive function aged 50–89 years: a cross-sectional study. Lancet Neurol. 13, 997–1005 (2014).
Boyle, P. A. et al. Much of late life cognitive decline is not due to common neurodegenerative pathologies. Ann. Neurol. 74, 478–489 (2013). This article shows that AD and age-related neuropathologies do not fully account for cognitive decline observed with ageing.
Murray, M. E. et al. Clinicopathologic and 11C-Pittsburgh compound B implications of Thal amyloid phase across the Alzheimer's disease spectrum. Brain http://dx.doi.org/10.1093/brain/awv050 (2015).
Sperling, R. A. et al. The A4 study: stopping AD before symptoms begin? Science Transl. Med. 6, 228fs213 (2014).
Villemagne, V. L. et al. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol. 12, 357–367 (2013). This work detected a long prodrome of amyloid build-up prior to the onset of cognitive symptoms of AD.
Dean, D. C. et al. Brain differences in infants at differential genetic risk for late-onset Alzheimer disease: a cross-sectional imaging study. JAMA Neurol. 71, 11–22 (2014).
Jansen, W. J. et al. Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA 313, 1924–1938 (2015).
Escott-Price, V. et al. Gene-wide analysis detects two new susceptibility genes for Alzheimer's disease. PLoS ONE 9, e94661 (2014).
Naj, A. C. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 43, 436–441 (2011).
Sabuncu, M. R. et al. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb. Cortex 22, 2653–2661 (2012).
Harris, S. E. et al. Polygenic risk for Alzheimer's disease is not associated with cognitive ability or cognitive aging in non-demented older people. J. Alzheimers Dis. 39, 565–574 (2014).
Ridge, P. G., Mukherjee, S., Crane, P. K., Kauwe, J. S. & Alzheimer's Disease Genetics Consortium. Alzheimer's disease: analyzing the missing heritability. PLoS ONE 8, e79771 (2013). This study considers the origin of the 'missing heritability' of AD, which appears when traditional additive models are used.
Cruchaga, C. et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature 505, 550–554 (2014).
Vardarajan, B. N. et al. Coding mutations in SORL1 and Alzheimer disease. Ann. Neurol. 77, 215–227 (2015).
Kobolt, D. et al. Exome-sequencing in a large dataset of late-onset families with Alzheimer's disease. Alzheimers Dement. 11, 359 (2015).
Farfel, J. et al. Relation of genomic variants for Alzheimer's disease dementia to common neuropathologies. Neurology (in the press) (2016).
Costanzo, M. et al. The genetic landscape of a cell. Science 327, 425–431 (2010).
He, X., Qian, W., Wang, Z., Li, Y. & Zhang, J. Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks. Nat. Genet. 42, 272–276 (2010).
Hu, T. et al. Characterizing genetic interactions in human disease association studies using statistical epistasis networks. BMC Bioinformatics 12, 364 (2011).
Pandey, A. et al. Epistasis network centrality analysis yields pathway replication across two GWAS cohorts for bipolar disorder. Transl. Psychiatry 2, e154 (2012).
Darabos, C. & Moore, J. H. in Epistasis 269–283 (Springer, 2015).
Moore, J. H. & Williams, S. M. Epistasis and its implications for personal genetics. Am. J. Hum. Genet. 85, 309–320 (2009).
Combarros, O., Cortina-Borja, M., Smith, A. D. & Lehmann, D. J. Epistasis in sporadic Alzheimer's disease. Neurobiol. Aging 30, 1333–1349 (2009).
Carrasquillo, M. M. et al. Replication of BIN1 association with Alzheimer's disease and evaluation of genetic interactions. J. Alzheimers Dis. 24, 751–758 (2011).
Gusareva, E. S. et al. Genome-wide association interaction analysis for Alzheimer's disease. Neurobiol. Aging 35, 2436–2443 (2014).
Bullock, J. M. et al. Discovery by the Epistasis Project of an epistatic interaction between the GSTM3 gene and the HHEX/IDE/KIF11 locus in the risk of Alzheimer's disease. Neurobiol. Aging 34, 1309.e1–1309.e7 (2013).
Meda, S. A. et al. Genetic interactions associated with 12-month atrophy in hippocampus and entorhinal cortex in Alzheimer's Disease Neuroimaging Initiative. Neurobiol. Aging 34, 1518.e9–1518.e18 (2013).
Hibar, D. P. et al. Genome-wide interaction analysis reveals replicated epistatic effects on brain structure. Neurobiol. Aging 36, S151–S158 (2015).
Zhong, Q. et al. Edgetic perturbation models of human inherited disorders. Mol. Syst. Biol. 5, 321 (2009). This work found that edge-specific (interface-specific) protein interactions, as opposed to all protein interactions, can be disrupted by many disease-related genetic variants.
Mosca, R. et al. dSysMap: exploring the edgetic role of disease mutations. Nat. Methods 12, 167–168 (2015).
Guo, Y. et al. Dissecting disease inheritance modes in a three-dimensional protein network challenges the 'guilt-by-association' principle. Am. J. Hum. Genet. 93, 78–89 (2013).
Sahni, N. et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660 (2015).
Vardarajan, B. N. et al. Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann. Neurol. 78, 487–498 (2015).
Jin, S. C. et al. Coding variants in TREM2 increase risk for Alzheimer's disease. Hum. Mol. Genet. 23, 5838–5846 (2014).
Feldman, I., Rzhetsky, A. & Vitkup, D. Network properties of genes harboring inherited disease mutations. Proc. Natl Acad. Sci. USA 105, 4323–4328 (2008).
Mitra, K., Carvunis, A.-R., Ramesh, S. K. & Ideker, T. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet. 14, 719–732 (2013).
Goh, K.-I. et al. The human disease network. Proc. Natl Acad. Sci. USA 104, 8685–8690 (2007). This study found widespread coherence in disease organization through the use of phenotypic similarity or gene signature overlap.
Köhler, S., Bauer, S., Horn, D. & Robinson, P. N. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 82, 949–958 (2008).
Raj, T. et al. Alzheimer disease susceptibility loci: evidence for a protein network under natural selection. Am. J. Hum. Genet. 90, 720–726 (2012).
O'Roak, B. J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012). This group used protein networks to show that, despite being seemingly sporadic, de novo mutations associated with autism are connected in a functionally related network that contains genes previously implicated in the disease.
Gulsuner, S. et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529 (2013).
Hormozdiari, F., Penn, O., Borenstein, E. & Eichler, E. E. The discovery of integrated gene networks for autism and related disorders. Genome Res. 25, 142–154 (2015).
Raj, T. et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014).
Pierson, E. et al. Sharing and specificity of co-expression networks across 35 human tissues. PLoS Comput. Biol. 11, e1004220 (2015).
Greene, C. S. et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Miller, J. A., Woltjer, R. L., Goodenbour, J. M., Horvath, S. & Geschwind, D. H. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Med. 5, 48 (2013).
Zhang, B. et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell 153, 707–720 (2013).
Neumann, H. & Daly, M. J. Variant TREM2 as risk factor for Alzheimer's disease. N. Engl. J. Med. 368, 182–184 (2013).
Hofree, M., Shen, J. P., Carter, H., Gross, A. & Ideker, T. Network-based stratification of tumor mutations. Nat. Methods 10, 1108–1115 (2013). In this study, protein interaction networks were used to organize mutations from individual patients.
Gligorijevic, V., Malod-Dognin, N. & Przulj, N. Patient-specific data fusion for cancer stratification and personalized treatment. Pac. Symp. Biocomput. 21, 321–332 (2016).
Leiserson, M. D. et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 47, 106–114 (2015).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008).
Tasaki, S. et al. Bayesian network reconstruction using systems genetics data: comparison of MCMC methods. Genetics 199, 973–989 (2015).
de la Fuente, A. From 'differential expression'to 'differential networking' — identification of dysfunctional regulatory networks in diseases. Trends Genet. 26, 326–333 (2010).
Rhinn, H. et al. Alternative α-synuclein transcript usage as a convergent mechanism in Parkinson's disease pathology. Nat. Commun. 3, 1084 (2012).
Amar, D., Safer, H. & Shamir, R. Dissection of regulatory networks that are altered in disease via differential co-expression. PLoS Comput. Biol. 9, e1002955 (2013).
Miller, J. A., Horvath, S. & Geschwind, D. H. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl Acad. Sci. USA 107, 12698–12703 (2010).
Schneider, J. A., Arvanitakis, Z., Leurgans, S. E. & Bennett, D. A. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann. Neurol. 66, 200–208 (2009).
Beecham, G. W. et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer's disease and related dementias. 10, e1004606 (2014).
Bennett, D. et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 66, 1837–1844 (2006).
Schneider, J. A., Arvanitakis, Z., Bang, W. & Bennett, D. A. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology 69, 2197–2204 (2007). Here, the researchers found that multiple types of pathology, in addition to classic AD pathology, account for clinical dementia in an older population.
De Jager, P. L. & Bennett, D. A. An inflection point in gene discovery efforts for neurodegenerative diseases: from syndromic diagnoses toward endophenotypes and the epigenome. JAMA Neurol. 70, 719–726 (2013).
Nelson, P. T. et al. ABCC9 gene polymorphism is associated with hippocampal sclerosis of aging pathology. Acta Neuropathol. 127, 825–843 (2014).
Hibar, D. P. et al. Common genetic variants influence human subcortical brain structures. Nature 520, 224–229 (2015).
Weiner, M. W. et al. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 8, S1–S68 (2012).
Schroeter, M. L., Stein, T., Maslowski, N. & Neumann, J. Neural correlates of Alzheimer's disease and mild cognitive impairment: a systematic and quantitative meta-analysis involving 1351 patients. Neuroimage 47, 1196–1206 (2009).
Scahill, R. I., Schott, J. M., Stevens, J. M., Rossor, M. N. & Fox, N. C. Mapping the evolution of regional atrophy in Alzheimer's disease: unbiased analysis of fluid-registered serial MRI. Proc. Natl Acad. Sci. USA 99, 4703–4707 (2002).
Betzel, R. F., Fukushima, M., He, Y., Zuo, X.-N. & Sporns, O. Dynamic fluctuations coincide with periods of high and low modularity in resting-state functional brain networks. Neuroimage 127, 287–297 (2016).
Fornito, A., Harrison, B. J., Zalesky, A. & Simons, J. S. Competitive and cooperative dynamics of large-scale brain functional networks supporting recollection. Proc. Natl Acad. Sci. USA 109, 12788–12793 (2012).
Sadaghiani, S., Poline, J.-B., Kleinschmidt, A. & D'Esposito, M. Ongoing dynamics in large-scale functional connectivity predict perception. Proc. Natl Acad. Sci. USA 112, 8463–8468 (2015).
Bassett, D. S., Meyer-Lindenberg, A., Achard, S., Duke, T. & Bullmore, E. Adaptive reconfiguration of fractal small-world human brain functional networks. Proc. Natl Acad. Sci. USA 103, 19518–19523 (2006).
van den Heuvel, M. P., Kahn, R. S., Goñi, J. & Sporns, O. High-cost, high-capacity backbone for global brain communication. Proc. Natl Acad. Sci. USA 109, 11372–11377 (2012).
van den Heuvel, M. P., Stam, C. J., Kahn, R. S. & Pol, H. E. H. Efficiency of functional brain networks and intellectual performance. J. Neurosci. 29, 7619–7624 (2009).
Gallos, L. K., Makse, H. A. & Sigman, M. A small world of weak ties provides optimal global integration of self-similar modules in functional brain networks. Proc. Natl Acad. Sci. USA 109, 2825–2830 (2012).
Watts, D. J. & Strogatz, S. H. Collective dynamics of 'small-world' networks. Nature 393, 440–442 (1998).
van den Heuvel, M. P. et al. Genetic control of functional brain network efficiency in children. Eur. Neuropsychopharmacol. 23, 19–23 (2013).
Bohlken, M. M. et al. Heritability of structural brain network topology: a DTI study of 156 twins. Hum. Brain Mapp. 35, 5295–5305 (2014).
Khazaee, A., Ebrahimzadeh, A. & Babajani-Feremi, A. Identifying patients with Alzheimer's disease using resting-state fMRI and graph theory. Clin. Neurophysiol. 126, 2132–2141 (2015).
Wang, J. et al. Disrupted functional brain connectome in individuals at risk for Alzheimer's disease. Biol. Psychiatry 73, 472–481 (2013).
Nir, T. M. et al. Connectivity network measures predict volumetric atrophy in mild cognitive impairment. Neurobiol. Aging 36, S113–S120 (2015).
Fischer, F. U., Wolf, D., Scheurich, A., Fellgiebel, A. & Alzheimer's Disease Neuroimaging Initiative. Altered whole-brain white matter networks in preclinical Alzheimer's disease. Neuroimage Clin. 8, 660–666 (2015).
Stam, C., Jones, B., Nolte, G., Breakspear, M. & Scheltens, P. Small-world networks and functional connectivity in Alzheimer's disease. Cereb. Cortex 17, 92–99 (2007). This study found initial evidence for conceptualization of widespread connectivity changes in AD as an altered balance of functional modularity and integration.
Lo, C.-Y. et al. Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer's disease. J. Neurosci. 30, 16876–16885 (2010).
Myers, N. et al. Within-patient correspondence of amyloid-β and intrinsic network connectivity in Alzheimer's disease. Brain 137, 2052–2064 (2014).
Frantzidis, C. A. et al. Functional disorganization of small-world brain networks in mild Alzheimer's disease and amnestic mild cognitive impairment: an EEG study using Relative Wavelet Entropy (RWE). Front. Aging Neurosci. 6, 224 (2014).
Stam, C. et al. Graph theoretical analysis of magnetoencephalographic functional connectivity in Alzheimer's disease. Brain 132, 213–224 (2009).
He, Y., Chen, Z. & Evans, A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J. Neurosci. 28, 4756–4766 (2008).
Langer, N. et al. Functional brain network efficiency predicts intelligence. Hum. Brain Mapp. 33, 1393–1406 (2012).
Liang, P., Wang, Z., Yang, Y., Jia, X. & Li, K. Functional disconnection and compensation in mild cognitive impairment: evidence from DLPFC connectivity using resting-state fMRI. PLoS ONE 6, e22153 (2011).
Gardini, S. et al. Increased functional connectivity in the default mode network in mild cognitive impairment: a maladaptive compensatory mechanism associated with poor semantic memory performance. J. Alzheimers Dis. 45, 457–470 (2015).
Crossley, N. A. et al. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain 137, 2382–2395 (2014).
Buckner, R. L. et al. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J. Neurosci. 29, 1860–1873 (2009).
de Haan, W., Mott, K., van Straaten, E. C., Scheltens, P. & Stam, C. J. Activity dependent degeneration explains hub vulnerability in Alzheimer's disease. PLoS Comput. Biol. 8, e1002582 (2012).
Seo, E. H. et al. Whole-brain functional networks in cognitively normal, mild cognitive impairment, and Alzheimer's disease. PLoS ONE 8, e53922 (2013).
Zamora-López, G., Zhou, C. & Kurths, J. Cortical hubs form a module for multisensory integration on top of the hierarchy of cortical networks. Front. Neuroinform. 4, 1 (2010).
de Reus, M. A. & van den Heuvel, M. P. Simulated rich club lesioning in brain networks: a scaffold for communication and integration? Front. Hum. Neurosci. 8, 647 (2014).
Wang, J. et al. Apolipoprotein E ε4 modulates functional brain connectome in Alzheimer's disease. Hum. Brain Mapp. 36, 1828–1846 (2015).
Dai, Z. et al. Identifying and mapping connectivity patterns of brain network hubs in Alzheimer's disease. Cereb. Cortex 25, 3723–3742 (2015).
Daianu, M. et al. Rich club analysis in the Alzheimer's disease connectome reveals a relatively undisturbed structural core network. Hum. Brain Mapp. 36, 3087–3103 (2015).
Daianu, M. et al. Disrupted rich club network in behavioral variant frontotemporal dementia and early-onset Alzheimer's disease. Hum. Brain Mapp. 37, 868–883 (2016).
Filippini, N. et al. Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. Proc. Natl Acad. Sci. USA 106, 7209–7214 (2009).
Sheline, Y. I. et al. APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Aβ42. J. Neurosci. 30, 17035–17040 (2010).
Patel, K. T. et al. Default mode network activity and white matter integrity in healthy middle-aged ApoE4 carriers. Brain Imaging Behav. 7, 60–67 (2013).
Goryawala, M. et al. Apolipoprotein-E4 (ApoE4) carriers show altered small-world properties in the default mode network of the brain. Biomed. Phys. Eng. Express 1, 015001 (2015).
Trachtenberg, A. J. et al. The effects of APOE on the functional architecture of the resting brain. Neuroimage 59, 565–572 (2012).
Shaw, P. et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 6, 494–500 (2007).
Jones, D. et al. Age-related changes in the default mode network are more advanced in Alzheimer disease. Neurology 77, 1524–1531 (2011).
Greicius, M. D., Srivastava, G., Reiss, A. L. & Menon, V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc. Natl Acad. Sci. USA 101, 4637–4642 (2004).
Langbaum, J. B. et al. Categorical and correlational analyses of baseline fluorodeoxyglucose positron emission tomography images from the Alzheimer's Disease Neuroimaging Initiative (ADNI). Neuroimage 45, 1107–1116 (2009).
Chhatwal, J. P. et al. Impaired default network functional connectivity in autosomal dominant Alzheimer disease. Neurology 81, 736–744 (2013).
Jahanshad, N. et al. in Multimodal Brain Image Analysis 29–40 (Springer, 2012).
Brier, M. R. et al. Functional connectivity and graph theory in preclinical Alzheimer's disease. Neurobiol. Aging 35, 757–768 (2014).
Tijms, B. M. et al. Single-subject grey matter graphs in Alzheimer's disease. PLoS ONE 8, e58921 (2013).
Sanz-Arigita, E. J. et al. Loss of 'small-world' networks in Alzheimer's disease: graph analysis of FMRI resting-state functional connectivity. PLoS ONE 5, e13788 (2010).
Tijms, B. M. et al. Gray matter network disruptions and amyloid beta in cognitively normal adults. Neurobiol. Aging 37, 154–160 (2016).
de Haan, W. et al. Functional neural network analysis in frontotemporal dementia and Alzheimer's disease using EEG and graph theory. BMC Neurosci. 10, 101 (2009).
Fischer, F. U., Wolf, D., Scheurich, A., Fellgiebel, A. & Alzheimer's Disease Neuroimaging Initiative. Altered whole-brain white matter networks in preclinical Alzheimer's disease. Neuroimage Clin. 8, 660–666 (2015).
Zhao, X. et al. Disrupted small-world brain networks in moderate Alzheimer's disease: a resting-state FMRI study. PLoS ONE 7, e33540 (2012).
Li, Y., Qin, Y., Chen, X. & Li, W. Exploring the functional brain network of Alzheimer's disease: based on the computational experiment. PLoS ONE 8, e73186 (2013).
Supekar, K., Menon, V., Rubin, D., Musen, M. & Greicius, M. D. Network analysis of intrinsic functional brain connectivity in Alzheimer's disease. PLoS Comput. Biol. 4, e1000100 (2008).
Yao, Z. et al. Abnormal cortical networks in mild cognitive impairment and Alzheimer's disease. PLoS Comput. Biol. 6, e1001006 (2010).
Honey, C. et al. Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl Acad. Sci. USA 106, 2035–2040 (2009).
Hosseini, S. H. & Kesler, S. R. Comparing connectivity pattern and small-world organization between structural correlation and resting-state networks in healthy adults. Neuroimage 78, 402–414 (2013).
Tagliazucchi, E. & Laufs, H. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron 82, 695–708 (2014).
Chang, C. et al. Association between heart rate variability and fluctuations in resting-state functional connectivity. Neuroimage 68, 93–104 (2013).
Airan, R. D. et al. Factors affecting characterization and localization of interindividual differences in functional connectivity using MRI. Hum. Brain Mapp. 37, 1986–1997 (2016).
Zalesky, A., Fornito, A., Cocchi, L., Gollo, L. L. & Breakspear, M. Time-resolved resting-state brain networks. Proc. Natl Acad. Sci. USA 111, 10341–10346 (2014).
Allen, E. A. et al. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 24, 663–676 (2014).
Hutchison, R. M. et al. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage 80, 360–378 (2013).
Rashid, B., Damaraju, E., Pearlson, G. D. & Calhoun, V. D. Dynamic connectivity states estimated from resting fMRI Identify differences among schizophrenia, bipolar disorder, and healthy control subjects. Front. Hum. Neurosci. 8, 897 (2014).
Kaiser, R. H. et al. Dynamic resting-state functional connectivity in major depression. Neuropsychopharmacology 41, 1822–1830 (2016).
Jones, D. T. et al. Non-stationarity in the 'resting brain's' modular architecture. PLoS ONE 7, e39731 (2012).
Peraza, L. R., Taylor, J.-P. & Kaiser, M. Divergent brain functional network alterations in dementia with Lewy bodies and Alzheimer's disease. Neurobiol. Aging 36, 2458–2467 (2015).
Sompolinsky, H. Computational neuroscience: beyond the local circuit. Curr. Opin. Neurobiol. http://dx.doi.org/10.1016/j.conb.2014.02.002 (2014).
Bhalla, U. S. Molecular computation in neurons: a modeling perspective. Curr. Opin. Neurobiol. 25, 31–37 (2014).
Linderman, J. J., Cilfone, N. A., Pienaar, E., Gong, C. & Kirschner, D. E. A multi-scale approach to designing therapeutics for tuberculosis. Integr. Biol. (Camb.) 7, 591–609 (2015).
Obiol-Pardo, C., Gomis-Tena, J., Sanz, F., Saiz, J. & Pastor, M. A multiscale simulation system for the prediction of drug-induced cardiotoxicity. J. Chem. Inf. Model. 51, 483–492 (2011).
Osborne, J. et al. A hybrid approach to multi-scale modelling of cancer. Phil. Trans. R. Soc. A Math. Phys. Eng. Sci. 368, 5013–5028 (2010).
Silva, J. R. et al. A multiscale model linking ion-channel molecular dynamics and electrostatics to the cardiac action potential. Proc. Natl Acad. Sci. USA 106, 11102–11106 (2009).
Barttfeld, P. et al. Signature of consciousness in the dynamics of resting-state brain activity. Proc. Natl Acad. Sci. USA 112, 887–892 (2015).
Spiros, A., Carr, R. & Geerts, H. Not all partial dopamine D2 receptor agonists are the same in treating schizophrenia. Exploring the effects of bifeprunox and aripiprazole using a computer model of a primate striatal dopaminergic synapse. Neuropsychiatr. Dis. Treat. 6, 589 (2010).
Anticevic, A. et al. NMDA receptor function in large-scale anticorrelated neural systems with implications for cognition and schizophrenia. Proc. Natl Acad. Sci. USA 109, 16720–16725 (2012).
Vattikuti, S. & Chow, C. C. A computational model for cerebral cortical dysfunction in autism spectrum disorders. Biol. Psychiatry 67, 672–678 (2010).
Foley, A. M., Ammar, Z. M., Lee, R. H. & Mitchell, C. S. Systematic review of the relationship between amyloid-β levels and measures of transgenic mouse cognitive deficit in Alzheimer's disease. J. Alzheimers Dis. 44, 787–795 (2015).
Franco, R. & Cedazo-Minguez, A. Successful therapies for Alzheimer's disease: why so many in animal models and none in humans. Front. Pharmacol. 5, 146 (2014).
Burns, T. C., Li, M. D., Mehta, S., Awad, A. J. & Morgan, A. A. Mouse models rarely mimic the transcriptome of human neurodegenerative diseases: a systematic bioinformatics-based critique of preclinical models. Eur. J. Pharmacol. 759, 101–117 (2015).
Przytycka, T. M., Singh, M. & Slonim, D. K. Toward the dynamic interactome: it's about time. Brief. Bioinform. 11, 15–29 (2010).
Ohyama, T. et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639 (2015).
Schadt, E. E., Buchanan, S., Brennand, K. J. & Merchant, K. M. Evolving toward a human-cell based and multiscale approach to drug discovery for CNS disorders. Front. Pharmacol. 5, 252 (2014).
Southern, J. et al. Multi-scale computational modelling in biology and physiology. Prog. Biophys. Mol. Biol. 96, 60–89 (2008).
Neymotin, S. et al. Calcium regulation of HCN channels supports persistent activity in a multiscale model of neocortex. Neuroscience 316, 344–366 (2016).
Rowan, M. S., Neymotin, S. A. & Lytton, W. W. Electrostimulation to reduce synaptic scaling driven progression of Alzheimer's disease. Front. Comput. Neurosci. 8, 39 (2014).
DeWoskin, D. et al. Distinct roles for GABA across multiple timescales in mammalian circadian timekeeping. Proc. Natl Acad. Sci. USA 112, E3911–E3919 (2015).
Myung, J. et al. GABA-mediated repulsive coupling between circadian clock neurons in the SCN encodes seasonal time. Proc. Natl Acad. Sci. USA 112, E3920–E3929 (2015).
Sanz Leon, P. et al. The Virtual Brain: a simulator of primate brain network dynamics. Front. Neuroinform. 7, 10 (2013).
Izhikevich, E. M. & Edelman, G. M. Large-scale model of mammalian thalamocortical systems. Proc. Natl Acad. Sci. USA 105, 3593–3598 (2008). This work showed that a highly simplified neuronal model could be connected realistically with other models to reproduce large-scale patterns of brain activation.
Eliasmith, C. & Trujillo, O. The use and abuse of large-scale brain models. Curr. Opin. Neurobiol. 25, 1–6 (2014).
Cabral, J. et al. Structural connectivity in schizophrenia and its impact on the dynamics of spontaneous functional networks. Chaos 23, 046111 (2013).
Chaudhuri, R., Knoblauch, K., Gariel, M.-A., Kennedy, H. & Wang, X.-J. A large-scale circuit mechanism for hierarchical dynamical processing in the primate cortex. Neuron 88, 419–431 (2015).
Nageswaran, J. M., Dutt, N., Krichmar, J. L., Nicolau, A. & Veidenbaum, A. V. A configurable simulation environment for the efficient simulation of large-scale spiking neural networks on graphics processors. Neural Netw. 22, 791–800 (2009).
Markram, H. et al. Reconstruction and simulation of neocortical microcircuitry. Cell 163, 456–492 (2015).
Potjans, T. C. & Diesmann, M. The cell-type specific cortical microcircuit: relating structure and activity in a full-scale spiking network model. Cereb. Cortex 24, 785–806 (2014).
Izhikevich, E. M. Simple model of spiking neurons. IEEE Trans. Neural Netw. 14, 1569–1572 (2003).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Oldoni, F. et al. Post-GWAS methodologies for localisation of functional non-coding variants: ANGPTL3. Atherosclerosis 246, 193–201 (2015).
Rajagopal, N. et al. High-throughput mapping of regulatory DNA. Nat. Biotechnol. 34, 167–174 (2016).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Maurano, M. T. et al. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 47, 1393–1401 (2015).
Akbarian, S. et al. The PsychENCODE project. Nat. Neurosci. 18, 1707–1712 (2015).
Gutenkunst, R. N. et al. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 3, e189 (2007).
Karr, J. R. et al. A whole-cell computational model predicts phenotype from genotype. Cell 150, 389–401 (2012).
Jolivet, R., Coggan, J. S., Allaman, I. & Magistretti, P. J. Multi-timescale modeling of activity-dependent metabolic coupling in the neuron-glia-vasculature ensemble. PLoS Comput. Biol. 11, e1004036–e1004036 (2015).
Li, W.-K., Hausknecht, M. J., Stone, P. & Mauk, M. D. Using a million cell simulation of the cerebellum: network scaling and task generality. Neural Netw. 47, 95–102 (2013).
Insel, T. R., Landis, S. C. & Collins, F. S. The NIH brain initiative. Science 340, 687–688 (2013).
Toga, A. W., Clark, K. A., Thompson, P. M., Shattuck, D. W. & Van Horn, J. D. Mapping the human connectome. Neurosurgery 71, 1 (2012).
Lichtman, J. W., Pfister, H. & Shavit, N. The big data challenges of connectomics. Nat. Neurosci. 17, 1448–1454 (2014).
O'Leary, T., Sutton, A. C. & Marder, E. Computational models in the age of large datasets. Curr. Opin. Neurobiol. 32, 87–94 (2015).
Rzhetsky, A., Foster, J. G., Foster, I. T. & Evans, J. A. Choosing experiments to accelerate collective discovery. Proc. Natl Acad. Sci. USA 112, 14569–14574 (2015). This work demonstrates that exploration of high risk, interdisciplinary research brings personal benefits and increases efficiency of scientific discovery.
Uzzi, B., Mukherjee, S., Stringer, M. & Jones, B. Atypical combinations and scientific impact. Science 342, 468–472 (2013).
Thiele, I. et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 31, 419–425 (2013).
Mostafavi, S., Ray, D., Warde-Farley, D., Grouios, C. & Morris, Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9, S4 (2008).
Muldoon, S. F., Soltesz, I. & Cossart, R. Spatially clustered neuronal assemblies comprise the microstructure of synchrony in chronically epileptic networks. Proc. Natl Acad. Sci. USA 110, 3567–3572 (2013).
Bonifazi, P. et al. GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Science 326, 1419–1424 (2009).
Takahashi, N., Sasaki, T., Matsumoto, W., Matsuki, N. & Ikegaya, Y. Circuit topology for synchronizing neurons in spontaneously active networks. Proc. Natl Acad. Sci. USA 107, 10244–10249 (2010).
Kasthuri, N. et al. Saturated reconstruction of a volume of neocortex. Cell 162, 648–661 (2015).
Dobrin, R. et al. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 10, R55 (2009).
Gaiteri, C., Guilloux, J.-P., Lewis, D. A. & Sibille, E. Altered gene synchrony suggests a combined hormone-mediated dysregulated state in major depression. PLoS ONE 5, e9970 (2010).
Ardlie, K. G. et al. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Croft, D. P., Krause, J. & James, R. Social networks in the guppy (Poecilia reticulata). Proc. Biol. Sci. 271, S516–S519 (2004).
Kramer, A. D., Guillory, J. E. & Hancock, J. T. Experimental evidence of massive-scale emotional contagion through social networks. Proc. Natl Acad. Sci. USA 111, 8788–8790 (2014).
Christakis, N. A. & Fowler, J. H. Social contagion theory: examining dynamic social networks and human behavior. Stat. Med. 32, 556–577 (2013).
Magger, O., Waldman, Y. Y., Ruppin, E. & Sharan, R. Enhancing the prioritization of disease-causing genes through tissue specific protein interaction networks. PLoS Comput. Biol. 8, e1002690 (2012).
Das, J. & Yu, H. HINT: high-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol. 6, 92 (2012).
Rolland, T. et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014).
Obayashi, T. et al. COXPRESdb: a database of coexpressed gene networks in mammals. Nucleic Acids Res. 36, D77–D82 (2008).
Debette, S. et al. Genome-wide studies of verbal declarative memory in nondemented older people: the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Biol. Psychiatry 77, 749–763 (2015).
De Jager, P. L. et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol. Aging 33, 1017.e1–1017.e15 (2012).
Bennett, D. A., Schneider, J. A., Tang, Y., Arnold, S. E. & Wilson, R. S. The effect of social networks on the relation between Alzheimer's disease pathology and level of cognitive function in old people: a longitudinal cohort study. Lancet Neurol. 5, 406–412 (2006).
Buchman, A. et al. Total daily physical activity and the risk of AD and cognitive decline in older adults. Neurology 78, 1323–1329 (2012).
Wilson, R. S. et al. Life-span cognitive activity, neuropathologic burden, and cognitive aging. Neurology 81, 314–321 (2013).
Wilson, R. S. et al. Participation in cognitively stimulating activities and risk of incident Alzheimer disease. JAMA 287, 742–748 (2002).
Alladi, S. et al. Bilingualism delays age at onset of dementia, independent of education and immigration status. Neurology 81, 1938–1944 (2013).
Stern, Y. et al. Influence of education and occupation on the incidence of Alzheimer's disease. JAMA 271, 1004–1010 (1994).
Hall, C. et al. Cognitive activities delay onset of memory decline in persons who develop dementia. Neurology 73, 356–361 (2009).
Stern, Y. What is cognitive reserve? Theory and research application of the reserve concept. J. Int. Neuropsychol. Soc. 8, 448–460 (2002).
Landau, S. M. et al. Association of lifetime cognitive engagement and low β-amyloid deposition. Arch. Neurol. 69, 623–629 (2012).
Bennett, D., Schneider, J., Wilson, R., Bienias, J. & Arnold, S. Education modifies the association of amyloid but not tangles with cognitive function. Neurology 65, 953–955 (2005).
Deary, I. J., Johnson, W. & Houlihan, L. M. Genetic foundations of human intelligence. Hum. Genet. 126, 215–232 (2009).
Davies, G. et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Mol. Psychiatry 20, 183–192 (2015).
Chibnik, L. B. et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann. Neurol. 69, 560–569 (2011).
Barral, S. et al. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology 78, 1464–1471 (2012).
Zhang, X. et al. Bridging integrator 1 (BIN1) genotype effects on working memory, hippocampal volume, and functional connectivity in young healthy individuals. Neuropsychopharmacology 40, 1794–1803 (2015).
Carrasquillo, M. M. et al. Late-onset Alzheimer's risk variants in memory decline, incident mild cognitive impairment, and Alzheimer's disease. Neurobiol. Aging 36, 60–67 (2015).
Hayden, K. M., Lutz, M. W., Kuchibhatla, M., Germain, C. & Plassman, B. L. Effect of APOE and CD33 on cognitive decline. PLoS ONE 10, e0130419 (2015).
Acknowledgements
This work was supported by NIH grants U01AG46152, R01AG36042, P30AG10161, RF1AG15819, R01AG17917, R01AG36836.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, made substantial contributions to discussions of the content, wrote the article, and reviewed and/or edited the manuscript before submission.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
PowerPoint slides
Glossary
- Genetic variants
-
Insertion, deletion or alternative coding of DNA
- LOAD
-
Late-onset Alzheimer disease (LOAD) is the most common form of the neurodegenerative disease, typically diagnosed clinically after the age of 65 years and definitively diagnosed postmortem. LOAD is associated with functionally diverse, weak genetic variants
- Multiscale models
-
Mathematical or conceptual models that couple processes that occur on a varying range of physical or temporal scales, which are typically studied in isolation from each other
- Amyloid hypothesis
-
Proposal according to which the root cause of Alzheimer disease is the accumulation of amyloid-β (Aβ), with nuances around sufficiency and form of amyloid-β responsible for pathogenesis
- Amyloid precursor protein
-
Amyloid precursor protein (APP) is cleaved to form amyloid-β peptides. Presenilin (PSEN1 and PSEN2) mutations promote the cleavage of APP into plaque-forming peptides
- Apolipoprotein E
-
Apolipoprotein E is a protein that physically interacts with amyloid-β (Aβ) and tau; its interaction with Aβ influences plaque aggregation. Dosage of the ε4 allele of the APOE gene is the strongest genetic risk factor for late-onset Alzheimer disease
- PPAR-γ
-
Peroxisome proliferator- activated receptor γ (PPAR-γ) is a component of a nuclear receptor complex that includes the retinoid X receptor. This complex is activated endogenously by fatty acids and leads to transcription of the apolipoprotein E gene, among other genes
- Epistasis
-
When a combination of two or more genetic variants have a greater effect on a phenotype than their linear combination would predict
- Pleiotropic effects
-
The contribution of a single gene, or variants of that gene, to two or more 'non-related' phenotypes
- Directed networks
-
Networks where elements are connected by links with a specific direction that represents an asymmetric temporal or transfer function
- Cognitive reserve
-
Tolerance and adaptation to neuropathology, in part attributed to genetic and lifestyle-associated factors such as education and social activity, and their neural correlates
- Small-world organization
-
Network structure in which most nodes are connected by a small number of hops, yet form relatively isolated (modular) clusters
- Clustering coefficient
-
Measure of modularity around a network node. The coefficient represents the number of connections among neighbours of a node divided by the maximum possible number of connections among those neighbours
Rights and permissions
About this article
Cite this article
Gaiteri, C., Mostafavi, S., Honey, C. et al. Genetic variants in Alzheimer disease — molecular and brain network approaches. Nat Rev Neurol 12, 413–427 (2016). https://doi.org/10.1038/nrneurol.2016.84
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneurol.2016.84
This article is cited by
-
Assessment of Alzheimer-related pathologies of dementia using machine learning feature selection
Alzheimer's Research & Therapy (2023)
-
Toward a Brain-Inspired Theory of Artificial Learning
Cognitive Computation (2023)
-
Early impairment of cortical circuit plasticity and connectivity in the 5XFAD Alzheimer’s disease mouse model
Translational Psychiatry (2022)
-
Omics-based biomarkers discovery for Alzheimer's disease
Cellular and Molecular Life Sciences (2022)
-
Unified AI framework to uncover deep interrelationships between gene expression and Alzheimer’s disease neuropathologies
Nature Communications (2021)