Key Points
-
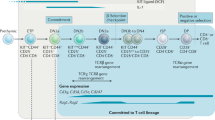
Zebrafish have an adaptive immune system that comprises both T and B cells. The molecular pathways that regulate the development and maturation of these lymphoid-cell populations have been highly conserved throughout evolution.
-
The zebrafish is a powerful genetic model for dissecting the pathways that regulate erythrocyte and leukocyte development.
-
Genetic screens have identified mutations that perturb normal thymic and T-cell development, most of which result in severe cranio-facial defects. This indicates that many of these mutations affect pharyngeal-arch development and, subsequently, that the thymic anlage fails to form in mutated embryos.
-
Although the main advantage of using the zebrafish model system lies in the ability to carry out forward-genetic screens, the zebrafish model also offers many other advantages for studying thymic and T-cell development.
-
Transgenic zebrafish have been created that have green fluorescent protein (GFP)-labelled T cells, which allow the visualization of thymocyte development and function in a living organism. In conjunction with cell transplantation, transgenic approaches have provided unique reagents for the study of T-cell homing and the assessment of haematopoietic stem-cell function.
-
Reverse-genetic approaches rely on the targeted inactivation of gene products in developing organisms. Because gene-knockout technology has yet to be developed in zebrafish, researchers can use either targeting induced local lesions in genomes (TILLING) or morpholino antisense oligonucleotides to disrupt gene function in developing embryos.
-
TILLING is a technique that uses N-ethyl-N-nitrosourea mutagenesis and the sequencing of genomic DNA to detect point mutations that are present in a gene of interest. This technology has been used successfully to disrupt recombination-activating gene 1 (rag1) gene function in zebrafish, which alters immune function in homozygous mutant individuals.
-
Morpholinos are chemically modified antisense oligonucleotides that can be microinjected into one-cell-stage embryos. Because of their long half-life, these molecules can disrupt gene function for up to 5 days.
-
The high-throughput sequencing of cDNA libraries in conjunction with whole-mount in situ hybridization has allowed researchers to carry out expression-based screens in zebrafish embryos, which provide new avenues for gene discovery.
-
Small-molecule-inhibitor screens have been used to identify water-soluble compounds that can be taken up by living zebrafish larvae and can restore mutant gene function in embryos. Because T-cell ablation can be monitored in rag2–GFP- and light-chain kinase (lck)–GFP-transgenic fish, and because dexamethasone treatment results in the loss of fluorescently labelled thymocytes, it might be possible to carry out small-molecule-inhibitor screens that are designed to identify drugs that selectively kill thymocytes, which might identify new immunosuppressive drugs.
Abstract
T-cell and thymic development are processes that have been highly conserved throughout vertebrate evolution. Mammals, birds, reptiles and fish share common molecular signalling pathways that regulate the development of the adaptive immune system. This Review article focuses on defining the similarities and differences between zebrafish and mammalian T-cell immunobiology, and it highlights the advantages of using the zebrafish as a genetic model to uncover mutations that affect T-cell and thymic development. Finally, we summarize the use of the zebrafish as a new model for assessing stem-cell function and for drug discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chitwood, B. G. & Chitwood, M. B. Introduction to Nematology (University Park, Baltimore, 1974).
Schulenburg, H., Kurz, C. L. & Ewbank, J. J. Evolution of the innate immune system: the worm perspective. Immunol. Rev. 198, 36–58 (2004).
Evans, C. J., Hartenstein, V. & Banerjee, U. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell 5, 673–690 (2003).
Appleby, M. W. & Ramsdell, F. A forward-genetic approach for analysis of the immune system. Nature Rev. Immunol. 3, 463–471 (2003).
Thisse, C. & Zon, L. I. Organogenesis: heart and blood formation from the zebrafish point of view. Science 295, 457–462 (2002).
Hansen, J. D. & Zapata, A. G. Lymphocyte development in fish and amphibians. Immunol. Rev. 166, 199–220 (1998).
Trede, N. S., Zapata, A. & Zon, L. I. Fishing for lymphoid genes. Trends Immunol. 22, 302–307 (2001).
Trede, N. S. & Zon, L. I. Development of T-cells during fish embryogenesis. Dev. Comp. Immunol. 22, 253–263 (1998).
Long, Q. et al. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development 124, 4105–4111 (1997).
Jessen, J. R., Willett, C. E. & Lin, S. Artificial chromosome transgenesis reveals long-distance negative regulation of rag1 in zebrafish. Nature Genet. 23, 15–16 (1999).
Jessen, J. R., Jessen, T. N., Vogel, S. S. & Lin, S. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis 29, 156–162 (2001).
Traver, D. et al. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nature Immunol. 4, 1238–1246 (2003). This paper describes the prospective isolation of specific blood-cell lineages in the kidney marrow of adult zebrafish through the use of FACS and GFP-transgenic technology. It also establishes methodologies for haematopoietic stem-cell transplantation into embryos.
Hsu, K. et al. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood 104, 1291–1297 (2004).
Langenau, D. M. et al. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl Acad. Sci. USA 101, 7369–7374 (2004). This work provides a comprehensive characterization of T- and B-cell lymphopoiesis in embryonic and adult zebrafish by comparing the expression of GFP in rag2–GFP - and lck–GFP -transgenic animals.
Streisinger, G., Walker, C., Dower, N., Knauber, D. & Singer, F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291, 293–296 (1981).
Driever, W. et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37–46 (1996).
Haffter, P. et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123, 1–36 (1996). References 16 and 17 are classic papers that describe the first ENU-based mutagenesis screens in zebrafish.
Poss, K. D., Nechiporuk, A., Hillam, A. M., Johnson, S. L. & Keating, M. T. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development 129, 5141–5149 (2002).
Peterson, R. T. et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nature Biotechnol. 22, 595–599 (2004).
Peterson, R. T., Link, B. A., Dowling, J. E. & Schreiber, S. L. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc. Natl Acad. Sci. USA 97, 12965–12969 (2000). Although small-molecule chemical screens using zebrafish were first described in reference 20, reference 19 describes the first use of small-molecule-inhibitor screens to identify compounds that restore mutant-gene function in living zebrafish embryos.
Laird, D. J., De Tomaso, A. W., Cooper, M. D. & Weissman, I. L. 50 million years of chordate evolution: seeking the origins of adaptive immunity. Proc. Natl Acad. Sci. USA 97, 6924–6926 (2000).
Du Pasquier, L., Zucchetti, I. & De Santis, R. Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol. Rev. 198, 233–248 (2004).
Greenhalgh, P. & Steiner, L. A. Recombination activating gene 1 (Rag1) in zebrafish and shark. Immunogenetics 41, 54–55 (1995).
Nam, B. H., Hirono, I. & Aoki, T. The four TCR genes of teleost fish: the cDNA and genomic DNA analysis of Japanese flounder (Paralichthys olivaceus) TCR α-, β-, γ-, and δ-chains. J. Immunol. 170, 3081–3090 (2003).
Partula, S., de Guerra, A., Fellah, J. S. & Charlemagne, J. Structure and diversity of the T cell antigen receptor β-chain in a teleost fish. J. Immunol. 155, 699–706 (1995).
Partula, S., Fellah, J. S., de Guerra, A. & Charlemagne, J. Characterization of cDNA of T-cell receptor β-chain in rainbow trout. C. R. Acad. Sci. III 317, 765–770 (1994).
Haire, R. N., Rast, J. P., Litman, R. T. & Litman, G. W. Characterization of three isotypes of immunoglobulin light chains and T-cell antigen receptor-α in zebrafish. Immunogenetics 51, 915–923 (2000).
Wilson, M. R. et al. T-cell receptors in channel catfish: structure and expression of TCR-α and -β genes. Mol. Immunol. 35, 545–557 (1998).
Wang, K. et al. Characterization of the Japanese pufferfish (Takifugu rubripes) T-cell receptor-α locus reveals a unique genomic organization. Immunogenetics 53, 31–42 (2001).
Wermenstam, N. E. & Pilstrom, L. T-cell antigen receptors in Atlantic cod (Gadus morhua L.): structure, organisation and expression of TCR-α and -β genes. Dev. Comp. Immunol. 25, 117–135 (2001).
Hordvik, I., Jacob, A. L., Charlemagne, J. & Endresen, C. Cloning of T-cell antigen receptor-β chain cDNAs from Atlantic salmon (Salmo salar). Immunogenetics 45, 9–14 (1996).
Schatz, D. G. Antigen receptor genes and the evolution of a recombinase. Semin. Immunol. 16, 245–256 (2004).
Willett, C. E., Kawasaki, H., Amemiya, C. T., Lin, S. & Steiner, L. A. Ikaros expression as a marker for lymphoid progenitors during zebrafish development. Dev. Dyn. 222, 694–698 (2001).
Park, C. I., Hirono, I., Enomoto, J., Nam, B. H. & Aoki, T. Cloning of Japanese flounder Paralichthys olivaceus CD3 cDNA and gene, and analysis of its expression. Immunogenetics 53, 130–135 (2001).
Park, C. I., Hirono, I. & Aoki, T. Molecular characterization of the Japanese flounder, Paralichthys olivaceus, CD3ε and evolution of the CD3 cluster. Dev. Comp. Immunol. 29, 123–133 (2005).
Suetake, H., Araki, K. & Suzuki, Y. Cloning, expression, and characterization of Fugu CD4, the first ectothermic animal CD4. Immunogenetics 56, 368–374 (2004).
Hansen, J. D. & Strassburger, P. Description of an ectothermic TCR coreceptor, CD8-α, in rainbow trout. J. Immunol. 164, 3132–3139 (2000).
Lam, S. H. et al. Morphologic transformation of the thymus in developing zebrafish. Dev. Dyn. 225, 87–94 (2002).
Willett, C. E., Cortes, A., Zuasti, A. & Zapata, A. G. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev. Dyn. 214, 323–336 (1999).
Willett, C. E., Zapata, A. G., Hopkins, N. & Steiner, L. A. Expression of zebrafish rag genes during early development identifies the thymus. Dev. Biol. 182, 331–341 (1997).
Danilova, N. et al. T cells and the thymus in developing zebrafish. Dev. Comp. Immunol. 28, 755–767 (2004).
Zon, L. I. Developmental biology of hematopoiesis. Blood 86, 2876–2891 (1995).
Johansson-Lindbom, B. et al. Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): requirement for GALT dendritic cells and adjuvant. J. Exp. Med. 198, 963–969 (2003).
Jahnsen, F. L., Farstad, I. N., Aanesen, J. P. & Brandtzaeg, P. Phenotypic distribution of T cells in human nasal mucosa differs from that in the gut. Am. J. Respir. Cell Mol. Biol. 18, 392–401 (1998).
Hirata, N., Takeuchi, K., Majima, Y. & Sakakura, Y. The Vδ1 T cell receptor repertoire in human nasal mucosa. Scand. J. Immunol. 52, 380–384 (2000).
Traver, D. et al. Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood 104, 1298–1305 (2004).
Lam, S. H., Chua, H. L., Gong, Z., Lam, T. J. & Sin, Y. M. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 28, 9–28 (2004).
Anderson, E. D. et al. Genetic immunization of rainbow trout (Oncorhynchus mykiss) against infectious hematopoietic necrosis virus. Mol. Mar. Biol. Biotechnol. 5, 114–122 (1996).
Lobb, C. & Clem, L. W. Fish lymphocytes differ in the expression of surface immunoglobulin. Dev. Comp. Immunol. 6, 473–479 (1982).
Lin, G. L., Ellsaesser, C. F., Clem, L. W. & Miller, N. W. Phorbol ester/calcium ionophore activate fish leukocytes and induce long-term cultures. Dev. Comp. Immunol. 16, 153–163 (1992).
Miller, N. et al. Functional and molecular characterization of teleost leukocytes. Immunol. Rev. 166, 187–197 (1998).
Miller, N. W., Deuter, A. & Clem, L. W. Phylogeny of lymphocyte heterogeneity: the cellular requirements for the mixed leucocyte reaction with channel catfish. Immunology 59, 123–128 (1986).
Vallejo, A. N., Miller, N. W. & Clem, L. W. Phylogeny of immune recognition: role of alloantigens in antigen presentation in channel catfish immune responses. Immunology 74, 165–168 (1991).
Saunders, H. L. & Magor, B. G. Cloning and expression of the AID gene in the channel catfish. Dev. Comp. Immunol. 28, 657–663 (2004).
Zhao, Y., Pan-Hammarstrom, Q., Zhao, Z. & Hammarstrom, L. Identification of the activation-induced cytidine deaminase gene from zebrafish: an evolutionary analysis. Dev. Comp. Immunol. 29, 61–71 (2005).
Stavnezer, J. & Amemiya, C. T. Evolution of isotype switching. Semin. Immunol. 16, 257–275 (2004).
Pancer, Z. et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430, 174–180 (2004).
Danilova, N. & Steiner, L. A. B cells develop in the zebrafish pancreas. Proc. Natl Acad. Sci. USA 99, 13711–13716 (2002).
Rolink, A., Haasner, D., Nishikawa, S. & Melchers, F. Changes in frequencies of clonable pre B cells during life in different lymphoid organs of mice. Blood 81, 2290–2300 (1993).
Brecher, G. et al. Self-renewal of the long-term repopulating stem cell. Proc. Natl Acad. Sci. USA 90, 6028–6031 (1993).
Perry, S. S., Pierce, L. J., Slayton, W. B. & Spangrude, G. J. Characterization of thymic progenitors in adult mouse bone marrow. J. Immunol. 170, 1877–1886 (2003).
Langenau, D. M. et al. Myc-induced T cell leukemia in transgenic zebrafish. Science 299, 887–890 (2003).
Nusslein-Volhard, C. & Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Papathanasiou, P. et al. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity 19, 131–144 (2003).
Nelms, K. A. & Goodnow, C. C. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity 15, 409–418 (2001).
Boehm, T., Bleul, C. C. & Schorpp, M. Genetic dissection of thymus development in mouse and zebrafish. Immunol. Rev. 195, 15–27 (2003).
Piotrowski, T. et al. The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in humans. Development 130, 5043–5052 (2003).
Popperl, H. et al. lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol. Cell 6, 255–267 (2000).
Reiter, J. F., Kikuchi, Y. & Stainier, D. Y. Multiple roles for Gata5 in zebrafish endoderm formation. Development 128, 125–135 (2001).
Ryan, A. K. et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J. Med. Genet. 34, 798–804 (1997).
Lindsay, E. A. et al. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410, 97–101 (2001).
Wittbrodt, J., Shima, A. & Schartl, M. Medaka: a model organism from the Far East. Nature Rev. Genet. 3, 53–64 (2002).
Iwanami, N. et al. Mutations affecting thymus organogenesis in medaka, Oryzias latipes. Mech. Dev. 121, 779–789 (2004).
Amsterdam, A. et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 13, 2713–2724 (1999). Work from the laboratory of Nancy Hopkins has established new methodologies for mutagen treatment and the rapid identification of disrupted gene products through the use of retroviral-based technologies. This is the seminal paper describing the development and use of retroviral gene disruption in the zebrafish.
Amsterdam, A. et al. Identification of 315 genes essential for early zebrafish development. Proc. Natl Acad. Sci. USA 101, 12792–12797 (2004).
Kawakami, K., Shima, A. & Kawakami, N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl Acad. Sci. USA 97, 11403–11408 (2000).
Parinov, S., Kondrichin, I., Korzh, V. & Emelyanov, A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 231, 449–459 (2004).
Kawakami, K. et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133–144 (2004).
Jansen, G., Hazendonk, E., Thijssen, K. L. & Plasterk, R. H. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nature Genet. 17, 119–121 (1997).
Bentley, A., MacLennan, B., Calvo, J. & Dearolf, C. R. Targeted recovery of mutations in Drosophila. Genetics 156, 1169–1173 (2000).
McCallum, C. M., Comai, L., Greene, E. A. & Henikoff, S. Targeted screening for induced mutations. Nature Biotechnol. 18, 455–457 (2000).
Perry, J. A. et al. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 131, 866–871 (2003).
Beier, D. R. Sequence-based analysis of mutagenized mice. Mamm. Genome 11, 594–597 (2000).
Coghill, E. L. et al. A gene-driven approach to the identification of ENU mutants in the mouse. Nature Genet. 30, 255–256 (2002).
Zan, Y. et al. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nature Biotechnol. 21, 645–651 (2003).
Hurlstone, A. F. et al. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature 425, 633–637 (2003).
Wienholds, E., Schulte-Merker, S., Walderich, B. & Plasterk, R. H. Target-selected inactivation of the zebrafish rag1 gene. Science 297, 99–102 (2002). TILLING approaches have been used in many organisms to identify point mutations that perturb gene function. This paper describes the first use of this reverse-genetic approach in the zebrafish.
Wienholds, E., Koudijs, M. J., van Eeden, F. J., Cuppen, E. & Plasterk, R. H. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nature Genet. 35, 217–218 (2003).
Wienholds, E. et al. Efficient target-selected mutagenesis in zebrafish. Genome Res. 13, 2700–2707 (2003).
Steinsvik, T. E., Gaarder, P. I., Aaberge, I. S. & Lovik, M. Engraftment and humoral immunity in SCID and RAG-2-deficient mice transplanted with human peripheral blood lymphocytes. Scand. J. Immunol. 42, 607–616 (1995).
Martin, A. et al. Engraftment of human lymphocytes and thyroid tissue into scid and rag2-deficient mice: absent progression of lymphocytic infiltration. J. Clin. Endocrinol. Metab. 79, 716–723 (1994).
Nasevicius, A. & Ekker, S. C. Effective targeted gene 'knockdown' in zebrafish. Nature Genet. 26, 216–220 (2000). This paper describes the first use of morpholino antisense technology in the zebrafish, providing a robust assay for assessing the loss of gene function in living zebrafish larvae.
Wang, J. H. et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5, 537–549 (1996).
Song, H. D. et al. Hematopoietic gene expression profile in zebrafish kidney marrow. Proc. Natl Acad. Sci. USA 101, 16240–16245 (2004).
Sinnhuber, R. O., Hendricks, J. D., Wales, J. H. & Putnam, G. B. Neoplasms in rainbow trout, a sensitive animal model for environmental carcinogenesis. Ann. NY Acad. Sci. 298, 389–408 (1978).
Hawkins, W. E., Overstreet, R. M., Fournie, J. W. & Walker, W. W. Development of aquarium fish models for environmental carcinogenesis: tumor induction in seven species. J. Appl. Toxicol. 5, 261–264 (1985).
Bunton, T. E. Experimental chemical carcinogenesis in fish. Toxicol. Pathol. 24, 603–618 (1996).
Spitsbergen, J. M. et al. Neoplasia in zebrafish (Danio rerio) treated with N-methyl-N'-nitro-N-nitrosoguanidine by three exposure routes at different developmental stages. Toxicol. Pathol. 28, 716–725 (2000).
Spitsbergen, J. M. et al. Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz(a)anthracene by two exposure routes at different developmental stages. Toxicol. Pathol. 28, 705–715 (2000).
Beckwith, L. G., Moore, J. L., Tsao-Wu, G. S., Harshbarger, J. C. & Cheng, K. C. Ethylnitrosourea induces neoplasia in zebrafish (Danio rerio). Lab. Invest. 80, 379–385 (2000).
Peterson, R. T., Mably, J. D., Chen, J. N. & Fishman, M. C. Convergence of distinct pathways to heart patterning revealed by the small molecule concentramide and the mutation heart-and-soul. Curr. Biol. 11, 1481–1491 (2001).
Zhong, T. P., Rosenberg, M., Mohideen, M. A., Weinstein, B. & Fishman, M. C. gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science 287, 1820–1824 (2000).
Langenau, D. M. et al. Suppression of apoptosis by bcl-2-overexpression in lymphoid cells of transgenic zebrafish. Blood 23 Dec 2004 (10.1182/blood-2004-08-3073).
Trede, N. S., Langenau, D. M., Traver, D., Look, A. T. & Zon, L. I. The use of zebrafish to understand immunity. Immunity 20, 367–379 (2004).
Acknowledgements
D.M.L. is the Edmond J. Safra Foundation–Irvington Institute Fellow. This work is supported by the National Institutes of Health (United States). L.I.Z. is also supported by the Howard Hughes Medical Institute (United States).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Gene
OMIM
T-cell acute lymphoblastic leukaemia
FURTHER INFORMATION
Glossary
- GYNOGENETIC
-
A term describing the production of offspring that have only maternally inherited chromosomes.
- ENU MUTAGENESIS
-
(N-ethyl-N-nitrosourea mutagenesis). Male fish are treated with ENU to induce point mutations in the spermatozoa and are then mated with female fish. The resulting progeny have heterozygous genetic lesions that are inherited from the father.
- BACTERIAL ARTIFICIAL CHROMOSOME
-
(BAC). A DNA construct that is used for cloning large genomic fragments into bacteria, which usually comprises 100–300 kilobases of genomic DNA.
- DIGEORGE SYNDROME
-
A rare congenital disease that often results in immune suppression, heart defects and cranio-facial defects. DiGeorge syndrome occurs because of a large deletion on chromosome 22, which results in the disruption of several genes from this genetic locus. Remarkably, the variation in symptoms might be related to the amount of genetic material that is lost by chromosomal deletion.
Rights and permissions
About this article
Cite this article
Langenau, D., Zon, L. The zebrafish: a new model of T-cell and thymic development. Nat Rev Immunol 5, 307–317 (2005). https://doi.org/10.1038/nri1590
Issue Date:
DOI: https://doi.org/10.1038/nri1590
This article is cited by
-
Cluster of differentiation antigens: essential roles in the identification of teleost fish T lymphocytes
Marine Life Science & Technology (2022)
-
In vivo impact of JAK3 A573V mutation revealed using zebrafish
Cellular and Molecular Life Sciences (2022)
-
Flow cytometry in the analysis of hematological parameters of tilapias: applications in environmental aquatic toxicology
Environmental Science and Pollution Research (2021)
-
Zebrafish cardiac regeneration—looking beyond cardiomyocytes to a complex microenvironment
Histochemistry and Cell Biology (2020)
-
Human macrophages survive and adopt activated genotypes in living zebrafish
Scientific Reports (2019)