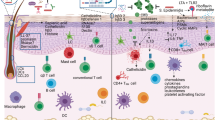
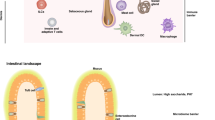
Key Points
-
The skin is a complex and dynamic ecosystem inhabited by many microorganisms.
-
The capacity of a given microorganism to trigger or promote disease is dependent on the state of immune activation of the host, the host's genetic predisposition and/or microorganism localization.
-
The skin microbiota can promote both innate and adaptive immunity to skin pathogens.
-
In many settings, optimal skin immunity is induced through networks of antigen-presenting cell subsets.
-
The skin is a large reservoir of tissue-resident memory T cells that can have an important role in protective immunity against pathogens.
-
Skin immune disorders in humans are associated with the enrichment of defined species of microorganisms.
Abstract
The skin is a complex and dynamic ecosystem that is inhabited by many microorganisms. Recent evidence highlights the profound reliance of the skin immune system on its resident microbiota for both host defence and tissue repair. This tissue is also a primary target for infections, which are in some cases caused by normal constituents of the microbiota. In the context of infections and genetic predispositions that are associated with barrier or regulatory network defects, microorganism-induced inflammatory cycles can contribute to the initiation and/or amplification of skin disorders. This Review will discuss some of our current understanding of skin–microbiota and skin–pathogen interactions in the context of homeostasis and diseases and highlight current gaps in our understanding of the skin immune ecosystem.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Buffie, C. G. & Pamer, E. G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 (2013).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Otto, M. Staphylococcus epidermidis — the 'accidental' pathogen. Nat. Rev. Microbiol. 7, 555–567 (2009).
Ibrahim, F., Khan, T. & Pujalte, G. G. Bacterial skin infections. Prim. Care 42, 485–499 (2015).
Roberts, N. & Horsley, V. Developing stratified epithelia: lessons from the epidermis and thymus. Wiley Interdiscip. Rev. Dev. Biol. 3, 389–402 (2014).
Pasparakis, M., Haase, I. & Nestle, F. O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 14, 289–301 (2014).
Tong, P. L. et al. The skin immune atlas: three-dimensional analysis of cutaneous leukocyte subsets by multiphoton microscopy. J. Invest. Dermatol. 135, 84–93 (2015).
Belkaid, Y. & Segre, J. A. Dialogue between skin microbiota and immunity. Science 346, 954–959 (2014).
Mueller, N. T., Bakacs, E., Combellick, J., Grigoryan, Z. & Dominguez-Bello, M. G. The infant microbiome development: mom matters. Trends Mol. Med. 21, 109–117 (2015).
PrabhuDas, M. et al. Challenges in infant immunity: implications for responses to infection and vaccines. Nat. Immunol. 12, 189–194 (2011).
Oh, J., Conlan, S., Polley, E. C., Segre, J. A. & Kong, H. H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 4, 77 (2012).
Si, J., Lee, S., Park, J. M., Sung, J. & Ko, G. Genetic associations and shared environmental effects on the skin microbiome of Korean twins. BMC Genomics 16, 992 (2015).
Scharschmidt, T. C. & Fischbach, M. A. What lives on our skin: ecology, genomics and therapeutic opportunities of the skin microbiome. Drug Discov. Today Dis. Mech. 10, pii: e83-e89 (2013).
Bouslimani, A. et al. Molecular cartography of the human skin surface in 3D. Proc. Natl Acad. Sci. USA 112, E2120–E2129 (2015).
Grice, E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009). References 15 and 16 reveal that the composition of the skin microbiota is site specific.
Song, S. J. et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2, e00458 (2013).
Lai, Y. et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 15, 1377–1382 (2009). This paper shows that a defined molecule produced by a commensal can indirectly promote repair.
Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012). This study reveals that the mirobiota can promote adaptive immunity in the skin.
Gallo, R. L. & Hooper, L. V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12, 503–516 (2012).
Nagy, I. et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect. 8, 2195–2205 (2006).
Chehoud, C. et al. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc. Natl Acad. Sci. USA 110, 15061–15066 (2013).
Verhulst, N. O. et al. Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS ONE 6, e28991 (2011).
Zhang, X., Crippen, T. L., Coates, C. J., Wood, T. K. & Tomberlin, J. K. Effect of quorum sensing by Staphylococcus epidermidis on the attraction response of female adult yellow fever mosquitoes, Aedes aegypti aegypti (Linnaeus) (Diptera: Culicidae), to a blood-feeding source. PLoS ONE 10, e0143950 (2015).
Naik, S. et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015).
Iwasaki, A. & Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353 (2015).
Hall, J. A. et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29, 637–649 (2008).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Volz, T. et al. Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL-10-producing dendritic cells and regulatory Tr1 cells. J. Invest. Dermatol. 134, 96–104 (2014).
Laborel-Preneron, E. et al. Effects of the Staphylococcus aureus and Staphylococcus epidermidis secretomes isolated from the skin microbiota of atopic children on CD4+ T cell activation. PLoS ONE 10, e0141067 (2015).
Skabytska, Y. et al. Cutaneous innate immune sensing of Toll-like receptor 2–6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity 41, 762–775 (2014).
Yockey, L. J. et al. The absence of a microbiota enhances TSLP expression in mice with defective skin barrier but does not affect the severity of their allergic inflammation. J. Invest. Dermatol. 133, 2714–2721 (2013).
Bantz, S. K., Zhu, Z. & Zheng, T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J. Clin. Cell Immunol. 5, pii:202 (2014).
Zhang, L. J. et al. Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 347, 67–71 (2015). This paper reveals a direct antimicrobial function from dermal adipocytes during skin infection.
Abtin, A. et al. Perivascular macrophages mediate neutrophil recruitment during bacterial skin infection. Nat. Immunol. 15, 45–53 (2014).
Otto, M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 17, 32–37 (2014).
Murphy, K. M. Transcriptional control of dendritic cell development. Adv. Immunol. 120, 239–267 (2013).
Malissen, B., Tamoutounour, S. & Henri, S. The origins and functions of dendritic cells and macrophages in the skin. Nat. Rev. Immunol. 14, 417–428 (2014).
Merad, M., Sathe, P., Helft, J., Miller, J. & Mortha, A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31, 563–604 (2013).
Hoeffel, G. et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med. 209, 1167–1181 (2012).
Haniffa, M., Bigley, V. & Collin, M. Human mononuclear phagocyte system reunited. Semin. Cell Dev. Biol. 41, 59–69 (2015).
Bedoui, S. et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat. Immunol. 10, 488–495 (2009). This paper shows that CD103+ DCs are the main skin migratory subtype that can cross-present viral and self antigens.
Haniffa, M. et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 37, 60–73 (2012).
Schlitzer, A. et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013).
Vander Lugt, B. et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat. Immunol. 15, 161–167 (2014).
Kolts, R. L., Maki, H. S., Kuehner, M. E., Roberts, R. C. & Sautter, R. D. Enhanced streptokinase-induced thrombolysis using heparin in a rabbit model. J. Invest. Surg. 2, 431–436 (1989).
Tussiwand, R. et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015).
Briseno, C. G., Murphy, T. L. & Murphy, K. M. Complementary diversification of dendritic cells and innate lymphoid cells. Curr. Opin. Immunol. 29, 69–78 (2014).
Scharschmidt, T. C. et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 (2015). This paper describes the characterization of FOXP3+ T reg accumulation in neonate skin and subsequent regulatory function in host–microbiota interaction.
Hansen, S. & Lehr, C. M. Transfollicular delivery takes root: the future for vaccine design? Expert Rev. Vaccines 13, 5–7 (2014).
Igyarto, B. Z. et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity 35, 260–272 (2011). This paper reveals specialization of skin-resident DCs during skin infection.
Kashem, S. W. et al. Candida albicans morphology and dendritic cell subsets determine T helper cell differentiation. Immunity 42, 356–366 (2015).
Kobayashi, T. et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 42, 756–766 (2015).
Kautz-Neu, K. et al. Langerhans cells are negative regulators of the anti-Leishmania response. J. Exp. Med. 208, 885–891 (2011).
Martinez-Lopez, M., Iborra, S., Conde-Garrosa, R. & Sancho, D. Batf3-dependent CD103+ dendritic cells are major producers of IL-12 that drive local Th1 immunity against Leishmania major infection in mice. Eur. J. Immunol. 45, 119–129 (2015).
Brewig, N. et al. Priming of CD8+ and CD4+ T cells in experimental leishmaniasis is initiated by different dendritic cell subtypes. J. Immunol. 182, 774–783 (2009).
Ng, L. G. et al. Migratory dermal dendritic cells act as rapid sensors of protozoan parasites. PLoS Pathog. 4, e1000222 (2008).
Seneschal, J., Clark, R. A., Gehad, A., Baecher-Allan, C. M. & Kupper, T. S. Human epidermal Langerhans cells maintain immune homeostasis in skin by activating skin resident regulatory T cells. Immunity 36, 873–884 (2012).
Akbari, M. et al. IRF4 in dendritic cells inhibits IL-12 production and controls Th1 immune responses against Leishmania major. J. Immunol. 192, 2271–2279 (2014).
Fabri, M. et al. Vitamin D is required for IFN-γ-mediated antimicrobial activity of human macrophages. Sci. Transl Med. 3, 104ra102 (2011).
Riol-Blanco, L. et al. Nociceptive sensory neurons drive interleukin-23-mediated psoriasiform skin inflammation. Nature 510, 157–161 (2014).
Kashem, S. W. et al. Nociceptive sensory fibers drive interleukin-23 production from CD301b+ dermal dendritic cells and drive protective cutaneous immunity. Immunity 43, 515–526 (2015).
Ribeiro-Gomes, F. L., Peters, N. C., Debrabant, A. & Sacks, D. L. Efficient capture of infected neutrophils by dendritic cells in the skin inhibits the early anti-leishmania response. PLoS Pathog. 8, e1002536 (2012).
Shen, W. et al. Adaptive immunity to murine skin commensals. Proc. Natl Acad. Sci. USA 111, E2977–E2986 (2014).
Scholz, F., Badgley, B. D., Sadowsky, M. J. & Kaplan, D. H. Immune mediated shaping of microflora community composition depends on barrier site. PLoS ONE 9, e84019 (2014).
Battaglia, M. et al. Induction of tolerance in type 1 diabetes via both CD4+CD25+ T regulatory cells and T regulatory type 1 cells. Diabetes 55, 1571–1580 (2006).
Lehman, H. Skin manifestations of primary immune deficiency. Clin. Rev. Allergy Immunol. 46, 112–119 (2014).
Rezaei, N., de Vries, E., Gambineri, E. & Haddad, E. in Stiehm's immune deficiencies. Chap. 1 (eds Sullivan, E. K. & Stiehm, E. R.) 3–59 (Academic, 2014).
Smeekens, S. P. et al. Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J. Innate Immun. 6, 253–262 (2014).
Kong, H. H. et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012).
van Rensburg, J. J. et al. The human skin microbiome associates with the outcome of and is influenced by bacterial infection. MBio 6, e01315–01315 (2015).
Belkaid, Y., Piccirilo. A. C., Mendez, S., Shevack, E. Sacks, D. L. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature 420, 502–507 (2002). This study is the first demonstration that T reg cells can mediate skin microbial persistence and maintenance of concomitant immunity.
Suffia, I. J., Reckling, S. K., Piccirillo, C. A., Goldszmid, R. S. & Belkaid, Y. Infected site-restricted Foxp3+ natural regulatory T cells are specific for microbial antigens. J. Exp. Med. 203, 777–788 (2006).
Dudda, J. C., Perdue, N., Bachtanian, E. & Campbell, D. J. Foxp3+ regulatory T cells maintain immune homeostasis in the skin. J. Exp. Med. 205, 1559–1565 (2008).
Godfrey, V. L., Wilkinson, J. E. & Russell, L. B. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am. J. Pathol. 138, 1379–1387 (1991).
Sanchez Rodriguez, R. et al. Memory regulatory T cells reside in human skin. J. Clin. Invest. 124, 1027–1036 (2014).
Belkaid, Y. & Tarbell, K. Regulatory T cells in the control of host-microorganism interactions*. Annu. Rev. Immunol. 27, 551–589 (2009).
Nosbaum, A. et al. Cutting edge: regulatory T cells facilitate cutaneous wound healing. J. Immunol. 196, 2010–2014 (2016).
Chiu, I. M. et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature 501, 52–57 (2013).
Witherden, D. A., Ramirez, K. & Havran, W. L. Multiple receptor-ligand interactions direct tissue-resident γδ T cell activation. Front. Immunol. 5, 602 (2014).
Laggner, U. et al. Identification of a novel proinflammatory human skin-homing Vγ9Vδ2 T cell subset with a potential role in psoriasis. J. Immunol. 187, 2783–2793 (2011).
Gray, E. E., Suzuki, K. & Cyster, J. G. Cutting edge: identification of a motile IL-17-producing γδ T cell population in the dermis. J. Immunol. 186, 6091–6095 (2011).
Sumaria, N. et al. Cutaneous immunosurveillance by self-renewing dermal γδ T cells. J. Exp. Med. 208, 505–518 (2011).
Gray, E. E. et al. Deficiency in IL-17-committed Vγ4+ γδ T cells in a spontaneous Sox13-mutant CD45.1+ congenic mouse substrain provides protection from dermatitis. Nat. Immunol. 14, 584–592 (2013).
Ramirez-Valle, F., Gray, E. E. & Cyster, J. G. Inflammation induces dermal Vγ4+ γδT17 memory- like cells that travel to distant skin and accelerate secondary IL-17-driven responses. Proc. Natl Acad. Sci. USA 112, 8046–8051 (2015).
Maher, B. M. et al. Nlrp-3-driven interleukin 17 production by γδT cells controls infection outcomes during Staphylococcus aureus surgical site infection. Infect. Immun. 81, 4478–4489 (2013).
Murphy, A. G. et al. Staphylococcus aureus infection of mice expands a population of memory γδ T cells that are protective against subsequent infection. J. Immunol. 192, 3697–3708 (2014).
Clark, R. A. et al. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 176, 4431–4439 (2006).
Carbone, F. R. Tissue-resident memory T cells and fixed immune surveillance in nonlymphoid organs. J. Immunol. 195, 17–22 (2015).
Schenkel, J. M. & Masopust, D. Tissue-resident memory T cells. Immunity 41, 886–897 (2014).
Mueller, S. N. & Mackay, L. K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16, 79–89 (2016).
Watanabe, R. et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl Med. 7, 279ra39 (2015).
Gebhardt, T. et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10, 524–530 (2009). This study uncovers for the first time a protective role for T RM cells.
Mueller, S. N., Gebhardt, T., Carbone, F. R. & Heath, W. R. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 31, 137–161 (2013).
Peters, N. C. et al. Chronic parasitic infection maintains high frequencies of short-lived Ly6C+CD4+ effector T cells that are required for protection against re-infection. PLoS Pathog. 10, e1004538 (2014).
Mackay, L. K. et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc. Natl Acad. Sci. USA 109, 7037–7042 (2012).
Shin, H. & Iwasaki, A. Generating protective immunity against genital herpes. Trends Immunol. 34, 487–494 (2013).
Owens, P., Han, G., Li, A. G. & Wang, X. J. The role of Smads in skin development. J. Invest. Dermatol. 128, 783–790 (2008).
Mackay, L. K. et al. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14, 1294–1301 (2013).
Suffia, I., Reckling, S., Salay, G. & Belkaid, Y. A role for CD103 in Treg retention at site of Leishmania major infection. J. Immunol. 174, 5444–5455 (2005).
Cyster, J. G. & Schwab, S. R. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu. Rev. Immunol. 30, 69–94 (2012).
Zaid, A. et al. Persistence of skin-resident memory T cells within an epidermal niche. Proc. Natl Acad. Sci. USA 111, 5307–5312 (2014).
Jiang, X. et al. Skin infection generates non-migratory memory CD8+ TRM cells providing global skin immunity. Nature 483, 227–231 (2012).
Gebhardt, T. et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature 477, 216–219 (2011).
Ariotti, S. et al. Tissue-resident memory CD8+ T cells continuously patrol skin epithelia to quickly recognize local antigen. Proc. Natl Acad. Sci. USA 109, 19739–19744 (2012).
Zhang, Q. et al. DOCK8 regulates lymphocyte shape integrity for skin antiviral immunity. J. Exp. Med. 211, 2549–2566 (2014).
Knickelbein, J. E. et al. Noncytotoxic lytic granule-mediated CD8+ T cell inhibition of HSV-1 reactivation from neuronal latency. Science 322, 268–271 (2008).
Muller, A. J. et al. CD4+ T cells rely on a cytokine gradient to control intracellular pathogens beyond sites of antigen presentation. Immunity 37, 147–157 (2012).
Schenkel, J. M. et al. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014).
Ariotti, S. et al. T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105 (2014). This paper reveals the bystander protective effect of T RM cells on skin immunity.
Crosby, E. J., Goldschmidt, M. H., Wherry, E. J. & Scott, P. Engagement of NKG2D on bystander memory CD8 T cells promotes increased immunopathology following Leishmania major infection. PLoS Pathog. 10, e1003970 (2014).
Leyden, J. J., Marples, R. R. & Kligman, A. M. Staphylococcus aureus in the lesions of atopic dermatitis. Br. J. Dermatol. 90, 525–530 (1974).
Gao, Z., Tseng, C. H., Strober, B. E., Pei, Z. & Blaser, M. J. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE 3, e2719 (2008).
Chalmers, R. J., O'Sullivan, T., Owen, C. M. & Griffiths, C. E. A systematic review of treatments for guttate psoriasis. Br. J. Dermatol. 145, 891–894 (2001).
Marples, R. R., Heaton, C. L. & Kligman, A. M. Staphylococcus aureus in psoriasis. Arch. Dermatol. 107, 568–570 (1973).
Nakamura, Y. et al. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 503, 397–401 (2013).
Niebuhr, M. et al. Staphylococcal α-toxin is a strong inducer of interleukin-17 in humans. Infect. Immun. 79, 1615–1622 (2011).
Dastgheyb, S. S. & Otto, M. Staphylococcal adaptation to diverse physiologic niches: an overview of transcriptomic and phenotypic changes in different biological environments. Future Microbiol. 10, 1981–1995 (2015).
Ichinohe, T. et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl Acad. Sci. USA 108, 5354–5359 (2011).
Grice, E. A. et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc. Natl Acad. Sci. USA 107, 14799–14804 (2010).
Fitz-Gibbon, S. et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol. 133, 2152–2160 (2013).
Kang, D., Shi, B., Erfe, M. C., Craft, N. & Li, H. Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci. Transl Med. 7, 293ra103 (2015).
Barker, J. N. et al. Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J. Investigative Dermatol. 127, 564–567 (2007).
Brown, S. J. et al. Prevalent and low-frequency null mutations in the filaggrin gene are associated with early-onset and persistent atopic eczema. J. Investigative Dermatol. 128, 1591–1594 (2008).
Natsuga, K., Cipolat, S. & Watt, F. M. Increased bacterial load and expression of antimicrobial peptides in skin of barrier-deficient mice with reduced cancer susceptibility. J. Invest. Dermatol. 136, 99–106 (2016).
Di Meglio, P., Villanova, F. & Nestle, F. O. Psoriasis. Cold Spring Harb. Perspect. Med. 4 (2014).
Hoste, E. et al. Innate sensing of microbial products promotes wound-induced skin cancer. Nat. Commun. 6, 5932 (2015).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Zeeuwen, P. L. et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 13, R101 (2012).
Hand, T. & Belkaid, Y. Microbial control of regulatory and effector T cell responses in the gut. Curr. Opin. Immunol. 22, 63–72 (2010).
Leung, D. Y., Walsh, P., Giorno, R. & Norris, D. A. A potential role for superantigens in the pathogenesis of psoriasis. J. Invest. Dermatol. 100, 225–228 (1993).
Reginald, K. et al. Immunoglobulin E antibody reactivity to bacterial antigens in atopic dermatitis patients. Clin. Exp. Allergy 41, 357–369 (2011).
Sims, J. E. & Smith, D. E. The IL-1 family: regulators of immunity. Nat. Rev. Immunol. 10, 89–102 (2010).
Donia, M. S. & Fischbach, M. A. Human microbiota. Small molecules from the human microbiota. Science 349, 1254766 (2015).
Medema, M. H. & Fischbach, M. A. Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648 (2015).
Marples, R. R., Downing, D. T. & Kligman, A. M. Control of free fatty acids in human surface lipids by Corynebacterium acnes. J. Invest. Dermatol. 56, 127–131 (1971).
Findley, K. et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370 (2013).
Oh, J. et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014).
Conlan, S. et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 13, R64 (2012).
Iwase, T. et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010).
Sacks, D. & Noben-Trauth, N. The immunology of susceptibility and resistance to Leishmania major in mice. Nat. Rev. Immunol. 2, 845–858 (2002).
Krishna, S. & Miller, L. S. Innate and adaptive immune responses against Staphylococcus aureus skin infections. Semin. Immunopathol. 34, 261–280 (2012).
Acknowledgements
The authors apologize to their colleagues for not having cited all papers relevant to this expanding field of research (and in particular older literature) because of space constraints and strict editorial limitation for reference number. This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases. S. T. was supported by an EMBO fellowship. The authors thank all members of the Belkaid laboratory for discussion and more particularly N. Bouladoux and J. Kehr for their editorial help with the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Mutualism
-
The relationship between two different species that live in close proximity and benefit from one another.
- Parasitism
-
The relationship between two different species that live in close proximity, in which one species (the parasite) benefits at the expense of the other (the host).
- Commensalism
-
The relationship between two different species, in which one species benefits from the other without affecting it.
- Antimicrobial peptides
-
(AMPs). Molecules that can rapidly kill or inactivate a diverse range of microorganisms, including Gram-negative and Gram-positive bacteria, fungi, viruses and parasites.
- Sebaceous glands
-
Small glands in the skin that secrete an oily matter (sebum) into the hair follicles to lubricate the skin and hair.
- Lipoteichoic acid
-
A major component of the cell wall of Gram-positive bacteria such as Staphylococcus epidermidis.
- Filaggrin
-
A filament-associated protein that binds to keratin fibres in epithelial cells and is important for maturation of skin epithelial cells into the corneocytes that form the outermost protective layer of human skin.
- Nociceptors
-
Sensory receptors on nerve cells that respond to potentially damaging stimuli by sending signals to the spinal cord and brain.
- Concomitant immunity
-
A paradoxical immune status in which resistance to reinfection coincides with the persistence of the original infection.
Rights and permissions
About this article
Cite this article
Belkaid, Y., Tamoutounour, S. The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol 16, 353–366 (2016). https://doi.org/10.1038/nri.2016.48
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri.2016.48