Key Points
-
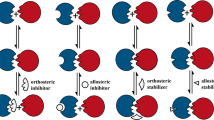
Protein–protein interactions (PPIs) are increasingly being targeted by drug discovery groups, and there exists great scope for therapeutic modulation of this target class in disease.
-
The array of structurally interacting elements through which proteins interact with one another is wide and resists clear-cut classification. However, broad divisions can be made by grouping interactions based upon the globular or peptidic nature of the proteins.
-
Some strategies for developing inhibitors against a given PPI may have more traction against certain classes of PPIs than others; for example, fragment-based drug discovery has shown particular promise in targeting bromodomains, as have peptide mimetics in mimicking β-strands.
-
We examine case studies representative of the various structural types of PPI and discuss the lessons learnt from each.
-
A summary of current status of inhibitors in clinical trials against different targets is presented.
Abstract
Protein–protein interactions (PPIs) are of pivotal importance in the regulation of biological systems and are consequently implicated in the development of disease states. Recent work has begun to show that, with the right tools, certain classes of PPI can yield to the efforts of medicinal chemists to develop inhibitors, and the first PPI inhibitors have reached clinical development. In this Review, we describe the research leading to these breakthroughs and highlight the existence of groups of structurally related PPIs within the PPI target class. For each of these groups, we use examples of successful discovery efforts to illustrate the research strategies that have proved most useful.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Petros, A. M. et al. Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J. Med. Chem. 49, 656–663 (2006).
Huggins, D. J., Marsh, M. & Payne, M. C. Thermodynamic properties of water molecules at a protein–protein interaction surface. J. Chem. Theory Comput. 7, 3514–3522 (2011).
Clackson, T. & Wells, J. A. A hot spot of binding energy in a hormone–receptor interface. Science 267, 383–386 (1995). A seminal paper presenting the crystal structure of human growth hormone and a domain of its receptor and systematically mutating interface residues to Ala.
Bogan, A. A. & Thorn, K. S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998). This paper examines the residue composition of hot spots at PPI interfaces, demonstrating an enrichment for certain amino acids. The importance of solvent occlusion around hot spot residues is also discussed.
Smith, M. C. & Gestwicki, J. E. Features of protein–protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Rev. Mol. Med. 14, e16 (2012).
Chakrabarti, P. & Janin, J. Dissecting protein–protein recognition sites. Proteins 47, 334–343 (2002).
Keskin, O., Ma, B. & Nussinov, R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 345, 1281–1294 (2005).
Ma, B., Elkayam, T., Wolfson, H. & Nussinov, R. Protein–protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl Acad. Sci. USA 100, 5772–5777 (2003).
Davis, F. P. & Sali, A. The overlap of small molecule and protein binding sites within families of protein structures. PLoS Comput. Biol. 6, e1000668 (2010).
Hu, Z., Ma, B., Wolfson, H. & Nussinov, R. Conservation of polar residues as hot spots at protein interfaces. Proteins 39, 331–342 (2000).
Sheinerman, F. B., Norel, R. & Honig, B. Electrostatic aspects of protein–protein interactions. Curr. Opin. Struct. Biol. 10, 153–159 (2000).
Rao, V. S., Srinivas, K., Sujini, G. N. & Kumar, G. N. Protein–protein interaction detection: methods and analysis. Int. J. Proteom. 2014, 147648 (2014).
Lehner, B. & Fraser, A. G. A first-draft human protein-interaction map. Genome Biol. 5, R63 (2004).
Zhang, Q. C. et al. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature 490, 556–560 (2012).
Winter, A. et al. Biophysical and computational fragment-based approaches to targeting protein–protein interactions: applications in structure-guided drug discovery. Q. Rev. Biophys. 45, 383–426 (2012).
Szklarczyk, D. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 (2011).
Higueruelo, A. P. et al. Atomic interactions and profile of small molecules disrupting protein–protein interfaces: the TIMBAL database. Chem. Biol. Drug Des. 74, 457–467 (2009).
Basse, M. J. et al. 2P2Idb: a structural database dedicated to orthosteric modulation of protein–protein interactions. Nucleic Acids Res. 41, D824–D827 (2013).
Morelli, X., Bourgeas, R. & Roche, P. Chemical and structural lessons from recent successes in protein–protein interaction inhibition (2P2I). Curr. Opin. Chem. Biol. 15, 475–481 (2011).
Bickerton, G. R., Higueruelo, A. P. & Blundell, T. L. Comprehensive, atomic-level characterization of structurally characterized protein–protein interactions: the PICCOLO database. BMC Bioinformat. 12, 313 (2011).
Jones, S. & Thornton, J. M. Principles of protein–protein interactions. Proc. Natl Acad. Sci. USA 93, 13–20 (1996). A classic paper reviewing PPIs, discussing concepts such as homo- and hetero-dimers as well as obligate and non-obligate complexes.
Miller, S. The structure of interfaces between subunits of dimeric and tetrameric proteins. Protein Eng. 3, 77–83 (1989).
Larsen, T. A., Olson, A. J. & Goodsell, D. S. Morphology of protein–protein interfaces. Structure 6, 421–427 (1998).
Keskin, O., Gursoy, A., Ma, B. & Nussinov, R. Principles of protein–protein interactions: what are the preferred ways for proteins to interact? Chem. Rev. 108, 1225–1244 (2008).
Arkin, M. R., Tang, Y. & Wells, J. A. Small-molecule inhibitors of protein–protein interactions: progressing toward the reality. Chem. Biol. 21, 1102–1114 (2014).
Blundell, T. L. et al. Structural biology and bioinformatics in drug design: opportunities and challenges for target identification and lead discovery. Phil. Trans. R. Soc. B 361, 413–423 (2006). This paper introduces the idea that protein structure can aid target selection, in particular the benefits of targeting a globular protein that orders a flexible peptide upon binding are proposed.
Wendt, M. D. Protein–protein interactions as drug targets. Top. Med. Chem. 8, 1–56 (2012).
Rickert, M., Wang, X., Boulanger, M. J., Goriatcheva, N. & Garcia, K. C. The structure of interleukin-2 complexed with its alpha receptor. Science 308, 1477–1480 (2005). This paper shows the crystal structure of IL-2 in complex with IL-2Rα, revealing the complex nature of the globular protein–globular protein interaction.
Jubb, H., Blundell, T. L. & Ascher, D. B. Flexibility and small pockets at protein–protein interfaces: new insights into druggability. Prog. Biophys. Mol. Biol. 119, 2–9 (2015).
Fletcher, S. & Prochownik, E. V. Small-molecule inhibitors of the Myc oncoprotein. Biochim. Biophys. Acta 1849, 525–543 (2015).
Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Follis, A. V., Hammoudeh, D. I., Wang, H., Prochownik, E. V. & Metallo, S. J. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem. Biol. 15, 1149–1155 (2008).
Hammoudeh, D. I., Follis, A. V., Prochownik, E. V. & Metallo, S. J. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 131, 7390–7401 (2009).
Pellegrini, L. et al. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 420, 287–293 (2002). The only published structure of human RAD51 published to date, revealing an interaction with a peptide derived from BRCA2.
Scott, D. E. et al. Small-molecule inhibitors that target protein–protein interactions in the RAD51 family of recombinases. ChemMedChem 10, 296–303 (2014).
Christ, F. et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 6, 442–448 (2010). A successful rational design of inhibitors binding to LEDGF (also known as p75) at a site bound by an inter-helix loop of viral HIV-1 integrase.
Fuller, J. C., Burgoyne, N. J. & Jackson, R. M. Predicting druggable binding sites at the protein–protein interface. Drug Discov. Today 14, 155–161 (2009).
Jochim, A. L. & Arora, P. S. Systematic analysis of helical protein interfaces reveals targets for synthetic inhibitors. ACS Chem. Biol. 5, 919–923 (2010).
Wells, J. A. & McClendon, C. L. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 450, 1001–1009 (2007).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Grasberger, B. L. et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J. Med. Chem. 48, 909–912 (2005).
Allen, J. G. et al. Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein–protein interaction. J. Med. Chem. 52, 7044–7053 (2009).
Blackburn, T. J. et al. Diaryl- and triaryl-pyrrole derivatives: inhibitors of the MDM2-p53 and MDMX-p53 protein–protein interactions. MedChemComm 4, 1297–1304 (2013).
Kenny, C. H. et al. Development of a fluorescence polarization assay to screen for inhibitors of the FtsZ/ZipA interaction. Anal. Biochem. 323, 224–233 (2003).
White, P. W. et al. Inhibition of human papillomavirus DNA replication by small molecule antagonists of the E1–E2 protein interaction. J. Biol. Chem. 278, 26765–26772 (2003).
Ferrari, S., Pellati, F. & Costi, M. P. in Disruption of Protein–Protein Interfaces (ed. Mangani, S.) 31–60 (Springer Berlin Heidelberg, 2013).
Hajduk, P. J. & Greer, J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug. Discov. 6, 211–219 (2007). An excellent review of fragment-based drug discovery with valuable historical insight.
Whittaker, M. Picking up the pieces with FBDD or FADD: invest early for future success. Drug Discov. Today 14, 623–624 (2009).
Blundell, T. L., Jhoti, H. & Abell, C. High-throughput crystallography for lead discovery in drug design. Nat.Rev. Drug. Discov. 1, 45–54 (2002).
Coyne, A. G., Scott, D. E. & Abell, C. Drugging challenging targets using fragment-based approaches. Curr. Opin. Chem. Biol. 14, 299–307 (2010).
Turnbull, A., Boyd, S. & Walse, B. Fragment-based drug discovery and protein–protein interactions. Res. Rep. Biochem. 4, 13–26 (2014).
Ciulli, A., Williams, G., Smith, A. G., Blundell, T. L. & Abell, C. Probing hot spots at protein-ligand binding sites: a fragment-based approach using biophysical methods. J. Med. Chem. 49, 4992–5000 (2006).
Scott, D. E. et al. Using a fragment-based approach to target protein–protein interactions. ChemBioChem 14, 332–342 (2013).
Lo, M. C. et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 332, 153–159 (2004).
Navratilova, I. & Hopkins, A. L. Emerging role of surface plasmon resonance in fragment-based drug discovery. Future. Med. Chem. 3, 1809–1820 (2011).
Lepre, C. A., Connolly, P. J. & Moore, J. M. in Drug Design (eds Merz, K. M. Jr., Ringe, D. & Reynolds, C. H.) 41–58 (Cambridge Univ. Press, 2010).
Davies, T. G. & Tickle, I. J. Fragment screening using X-ray crystallography. Top. Curr. Chem. 317, 33–59 (2012).
Turnbull, W. B. & Daranas, A. H. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 125, 14859–14866 (2003).
Erlanson, D. A. et al. Site-directed ligand discovery. Proc. Natl Acad. Sci. USA 97, 9367–9372 (2000). The first report of covalent 'tethering' as a technique for hit discovery and lead development against challenging drug discovery targets.
Morley, A. D. et al. Fragment-based hit identification: thinking in 3D. Drug Discov. Today 18, 1221–1227 (2013).
Van Molle, I. et al. Dissecting fragment-based lead discovery at the von Hippel-Lindau protein:hypoxia inducible factor 1α protein–protein interface. Chem. Biol. 19, 1300–1312 (2012).
Jhoti, H., Williams, G., Rees, D. C. & Murray, C. W. The 'rule of three' for fragment-based drug discovery: where are we now? Nat. Rev. Drug. Discov. 12, 644–645 (2013).
Fry, D. C. et al. Deconstruction of a nutlin: dissecting the binding determinants of a potent protein–protein interaction inhibitor. ACS Med. Chem. Lett. 4, 660–665 (2013).
Barelier, S., Pons, J., Marcillat, O., Lancelin, J.-M. & Krimm, I. Fragment-based deconstruction of Bcl-xL inhibitors. J. Med. Chem. 53, 2577–2588 (2010).
Peat, T. S. et al. Small molecule inhibitors of the LEDGF site of human immunodeficiency virus integrase identified by fragment screening and structure based design. PLoS ONE 7, e40147 (2012).
Ichihara, O., Barker, J., Law, R. J. & Whittaker, M. Compound design by fragment-linking. Mol. Inform. 30, 298–306 (2011).
de Vega, M. J., Martin-Martinez, M. & Gonzalez-Muniz, R. Modulation of protein–protein interactions by stabilizing/mimicking protein secondary structure elements. Curr. Top. Med. Chem. 7, 33–62 (2007).
Craik, D. J., Fairlie, D. P., Liras, S. & Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 81, 136–147 (2013).
Henchey, L. K., Jochim, A. L. & Arora, P. S. Contemporary strategies for the stabilization of peptides in the α-helical conformation. Curr. Opin. Chem. Biol. 12, 692–697 (2008).
Walensky, L. D. & Bird, G. H. Hydrocarbon-stapled peptides: principles, practice, and progress. J. Med. Chem. 57, 6275–6288 (2014). A review of the concept of 'stapled peptides' — hydrocarbon chains holding peptides into a fixed conformation.
Chu, Q. et al. Towards understanding cell penetration by stapled peptides. Med. Chem. Commun. 6, 111–119 (2014).
Kritzer, J. A. Stapled peptides: magic bullets in nature's arsenal. Nat. Chem. Biol. 6, 566–567 (2010).
Liu, J., Wang, D., Zheng, Q., Lu, M. & Arora, P. S. Atomic structure of a short α-helix stabilized by a main chain hydrogen-bond surrogate. J. Am. Chem. Soc. 130, 4334–4337 (2008).
Seebach, D. & Gardiner, J. β-peptidic peptidomimetics. Acc. Chem. Res. 41, 1366–1375 (2008).
Sadowsky, J. D. et al. Chimeric (α/β + α)-peptide ligands for the BH3-recognition cleft of Bcl-XL: critical role of the molecular scaffold in protein surface recognition. J. Am. Chem. Soc. 127, 11966–11968 (2005). An investigation of both α- and β-amino acid peptides, and hybrids of both, as inhibitors of the BH3–BCL-X L interaction.
Fletcher, S. & Hamilton, A. D. Protein surface recognition and proteomimetics: mimics of protein surface structure and function. Curr. Opin. Chem. Biol. 9, 632–638 (2005).
Chen, L. et al. p53 α-helix mimetics antagonize p53/MDM2 interaction and activate p53. Mol. Cancer Ther. 4, 1019–1025 (2005).
Davis, J. M., Truong, A. & Hamilton, A. D. Synthesis of a 2,3';6',3''-terpyridine scaffold as an α-helix mimetic. Org. Lett. 7, 5405–5408 (2005).
Shaginian, A. et al. Design, synthesis, and evaluation of an α-helix mimetic library targeting protein–protein interactions. J. Am. Chem. Soc. 131, 5564–5572 (2009). In this study a 400-member triaryl amide library, designed as α-helix mimetics, was screened against the MDM2–p53 PPI.
Bayly, A. R., White, A. J. P. & Spivey, A. C. Design and synthesis of a prototype scaffold for five-residue α-helix mimetics. Eur. J. Org. Chem. 25, 5566–5569 (2013).
Wei, C. Q., Li, B., Guo, R., Yang, D. & Burke, T. R. Jr. Development of a phosphatase-stable phosphotyrosyl mimetic suitably protected for the synthesis of high-affinity Grb2 SH2 domain-binding ligands. Bioorg. Med. Chem. Lett. 12, 2781–2784 (2002).
Wei, C. Q. et al. Macrocyclization in the design of Grb2 SH2 domain-binding ligands exhibiting high potency in whole-cell systems. J. Med. Chem. 46, 244–254 (2003).
Loughlin, W. A., Tyndall, J. D. A., Glenn, M. P., Hill, T. A. & Fairlie, D. P. Beta-strand mimetics. Chem. Rev. 110, 32–69 (2010).
Rognan, D. Rational design of protein–protein interaction inhibitors. Med. Chem. Commun. 6, 51–60 (2015).
Sugaya, N. & Furuya, T. Dr. PIAS: an integrative system for assessing the druggability of protein–protein interactions. BMC Bioinformatics 12, 50 (2011).
Walter, P., Metzger, J., Thiel, C. & Helms, V. Predicting where small molecules bind at protein–protein interfaces. PLoS ONE 8, e58583 (2013).
Meireles, L. M., Domling, A. S. & Camacho, C. J. ANCHOR: a web server and database for analysis of protein–protein interaction binding pockets for drug discovery. Nucleic Acids Res. 38, W407–W411 (2010).
Bienstock, R. J. Computational drug design targeting protein–protein interactions. Curr. Pharm. Des. 18, 1240–1254 (2012).
Grimme, D., González-ruiz, D. & Gohlke, H. in Physico-Chemical and Computational Approaches to Drug Discovery (eds Luque, J. & Barril, X.) 319–359 (2012).
Falchi, F., Caporuscio, F. & Recanatini, M. Structure-based design of small-molecule protein–protein interaction modulators: the story so far. Future Med. Chem. 6, 343–357 (2014).
Johnson, D. K. & Karanicolas, J. Druggable protein interaction sites are more predisposed to surface pocket formation than the rest of the protein surface. PLoS Comput. Biol. 9, e1002951 (2013).
Hajduk, P. J., Huth, J. R. & Fesik, S. W. Druggability indices for protein targets derived from NMR-based screening data. J. Med. Chem. 48, 2518–2525 (2005).
Brown, S. P. & Hajduk, P. J. Effects of conformational dynamics on predicted protein druggability. ChemMedChem 1, 70–72 (2006).
Tan, Y. S., Spring, D. R., Abell, C. & Verma, C. The use of chlorobenzene as a probe molecule in molecular dynamics simulations. J. Chem. Inf. Model. 54, 1821–1827 (2014).
Sledz, P. et al. From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1. Angew. Chem. Int. Ed. Engl. 50, 4003–4006 (2011).
Vogler, M., Dinsdale, D., Dyer, M. J. & Cohen, G. M. Bcl-2 inhibitors: small molecules with a big impact on cancer therapy. Cell Death Differ. 16, 360–367 (2009).
Souers, A. J. et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208 (2013).
Liu, X. Q., Dai, S. D., Zhu, Y. N., Marrack, P. & Kappler, J. W. The structure of a Bcl-xL/Bim fragment complex: implications for bim function. Immunity 19, 341–352 (2003).
Oberstein, A., Jeffrey, P. D. & Shi, Y. G. Crystal structure of the Bcl-X-L–beclin 1 peptide complex — beclin 1 is a novel BH3-only protein. J. Biol. Chem. 282, 13123–13132 (2007).
Lee, E. F. et al. Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J. Biol. Chem. 284, 30508–30517 (2009).
Petros, A. M. et al. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 9, 2528–2534 (2000).
Lee, E. F. et al. Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 14, 1711–1713 (2007).
Lee, E. F. et al. High-resolution structural characterization of a helical α/β-peptide foldamer bound to the anti-apoptotic protein Bcl-xL . Angew. Chem. Int. Ed. Engl. 48, 4318–4322 (2009).
Sattler, M. et al. Structure of Bcl-xL–Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986 (1997).
Wang, J. L. et al. Structure-based discovery of an organic compound that binds Bcl-2 protein and induces apoptosis of tumor cells. Proc. Natl Acad. Sci. USA 97, 7124–7129 (2000).
Enyedy, I. J. et al. Discovery of small-molecule inhibitors of Bcl-2 through structure-based computer screening. J. Med. Chem. 44, 4313–4324 (2001).
Lugovskoy, A. A. et al. A novel approach for characterizing protein ligand complexes: molecular basis for specificity of small-molecule Bcl-2 inhibitors. J. Am. Chem. Soc. 124, 1234–1240 (2002).
Mukherjee, P., Desai, P., Zhou, Y. D. & Avery, M. Targeting the BH3 domain mediated protein–protein interaction of Bcl-xL through virtual screening. J. Chem. Inf. Model. 50, 906–923 (2010).
Real, P. J. et al. Breast cancer cells can evade apoptosis-mediated selective killing by a novel small molecule inhibitor of Bcl-2. Cancer Res. 64, 7947–7953 (2004).
Wang, G. et al. Structure-based design of potent small-molecule inhibitors of anti-apoptotic Bcl-2 proteins. J. Med. Chem. 49, 6139–6142 (2006).
Zhou, H. et al. Design of Bcl-2 and Bcl-xL inhibitors with subnanomolar binding affinities based upon a new scaffold. J. Med. Chem. 55, 4664–4682 (2012).
Biros, S. M. et al. Heterocyclic α-helix mimetics for targeting protein–protein interactions. Bioorg. Med. Chem. Lett. 17, 4641–4645 (2007).
Yin, H. & Hamilton, A. D. Terephthalamide derivatives as mimetics of the helical region of Bak peptide target Bcl-xL protein. Bioorg. Med. Chem. Lett. 14, 1375–1379 (2004).
Antuch, W. et al. Design and modular parallel synthesis of a MCR derived α-helix mimetic protein–protein interaction inhibitor scaffold. Bioorg. Med. Chem. Lett. 16, 1740–1743 (2006).
Walensky, L. D. et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305, 1466–1470 (2004).
Stewart, M. L., Fire, E., Keating, A. E. & Walensky, L. D. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 6, 595–601 (2010).
Perez, H. L. et al. Identification of a phenylacylsulfonamide series of dual Bcl-2/Bcl-xL antagonists. Bioorg. Med. Chem. Lett. 22, 3946–3950 (2012).
Degterev, A. et al. Identification of small-molecule inhibitors of interaction between the BH3 domain and Bcl-xL. Nat. Cell Biol. 3, 173–182 (2001).
Tse, C. et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 68, 3421–3428 (2008).
Sleebs, B. E. et al. Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity. J. Med. Chem. 54, 1914–1926 (2011).
Tanaka, Y. et al. Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins. J. Med. Chem. 56, 9635–9645 (2013).
Mahon, A. B., Miller, S. E., Joy, S. T. & Arora, P. S. Rational design strategies for developing synthetic inhibitors of helical protein interfaces. Top. Med. Chem. 8, 197–230 (2012).
Zhao, Y., Aguilar, A., Bernard, D. & Wang, S. Small-molecule inhibitors of the MDM2-p53 protein–protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment. J. Med. Chem. 58, 1038–1052 (2015).
Srinivasula, S. M. et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410, 112–116 (2001).
Huang, Y., Rich, R. L., Myszka, D. G. & Wu, H. Requirement of both the second and third BIR domains for the relief of X-linked inhibitor of apoptosis protein (XIAP)-mediated caspase inhibition by Smac. J. Biol. Chem. 278, 49517–49522 (2003).
Sun, H. Y. et al. Structure-based design of potent, conformationally constrained Smac mimetics. J. Am. Chem. Soc. 126, 16686–16687 (2004).
Oost, T. K. et al. Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J. Med. Chem. 47, 4417–4426 (2004).
Cai, Q. et al. A potent and orally active antagonist (SM-406/AT-406) of multiple inhibitor of apoptosis proteins (IAPs) in clinical development for cancer treatment. J. Med. Chem. 54, 2714–2726 (2011).
Cossu, F. et al. Designing Smac-mimetics as antagonists of XIAP, cIAP1, and cIAP2. Biochem. Biophys. Res. Commun. 378, 162–167 (2009).
Monfardini, I. et al. Screening multicomponent reactions for X-linked inhibitor of apoptosis-baculoviral inhibitor of apoptosis protein repeats domain binder. J. Med. Chem. 54, 890–900 (2011).
Huang, J. W. et al. Fragment-based design of small molecule X-linked inhibitor of apoptosis protein inhibitors. J. Med. Chem. 51, 7111–7118 (2008).
Sun, H. et al. Design, synthesis, and characterization of a potent, nonpeptide, cell-permeable, bivalent Smac mimetic that concurrently targets both the BIR2 and BIR3 domains in XIAP. J. Am. Chem. Soc. 129, 15279–15294 (2007).
Wu, H., Tschopp, J. & Lin, S. C. Smac mimetics and TNFα: a dangerous liaison? Cell 131, 655–658 (2007).
Kester, R. F. et al. Optimization of benzodiazepinones as selective inhibitors of the X-linked inhibitor of apoptosis protein (XIAP) second baculovirus IAP repeat (BIR2) domain. J. Med. Chem. 56, 7788–7803 (2013).
Donnell, A. F. et al. Benzazepinones and benzoxazepinones as antagonists of inhibitor of apoptosis proteins (IAPs) selective for the second baculovirus IAP repeat (BIR2) domain. J. Med. Chem. 56, 7772–7787 (2013).
Padmanabhan, B. et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21, 689–700 (2006).
Tong, K. I. et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 27, 7511–7521 (2007). This paper proposes a hinge and latch model to explain how the KEAP1–NRF2 system senses and responds to oxidative and electrophilic stress.
Lo, S. C., Li, X., Henzl, M. T., Beamer, L. J. & Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 25, 3605–3617 (2006).
Chen, Y., Inoyama, D., Kong, A. N., Beamer, L. J. & Hu, L. Kinetic analyses of Keap1–Nrf2 interaction and determination of the minimal Nrf2 peptide sequence required for Keap1 binding using surface plasmon resonance. Chem. Biol. Drug Des. 78, 1014–1021 (2011).
Hancock, R. et al. Peptide inhibitors of the Keap1-Nrf2 protein–protein interaction. Free Radic. Biol. Med. 52, 444–451 (2012).
Hancock, R., Schaap, M., Pfister, H. & Wells, G. Peptide inhibitors of the Keap1–Nrf2 protein–protein interaction with improved binding and cellular activity. Org. Biomol. Chem. 11, 3553–3557 (2013).
Hu, L. et al. Discovery of a small-molecule inhibitor and cellular probe of Keap1–Nrf2 protein–protein interaction. Bioorg. Med. Chem. Lett. 23, 3039–3043 (2013).
Marcotte, D. et al. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorg. Med.Chem. 21, 4011–4019 (2013).
Jiang, Z. Y. et al. Discovery of potent Keap1–Nrf2 protein–protein interaction inhibitor based on molecular binding determinants analysis. J. Med. Chem. 57, 2736–2745 (2014). In this study a potent inhibitor of the KEAP1–NRF2 interaction was designed based upon a reported ligand of KEAP1 and a consideration of hot spot residues from the NRF2 peptide ligand.
Sun, H. P. et al. Novel protein–protein interaction inhibitor of Nrf2–Keap1 discovered by structure-based virtual screening. MedChemComm 5, 93–98 (2014).
Zhuang, C., Narayanapillai, S., Zhang, W., Sham, Y. Y. & Xing, C. Rapid identification of Keap1–Nrf2 small-molecule inhibitors through structure-based virtual screening and hit-based substructure search. J. Med.Chem. 57, 1121–1126 (2014).
Ducki, S. & Bennett, E. Protein–protein interactions: recent progress in the development of selective PDZ inhibitors. Curr. Chem. Biol. 3, 1–13 (2009).
Cox, D., Brennan, M. & Moran, N. Integrins as therapeutic targets: lessons and opportunities. Nat.Rev. Drug. Discov. 9, 804–820 (2010).
Patel, A., Dharmarajan, V. & Cosgrove, M. S. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J. Biol. Chem. 283, 32158–32161 (2008).
Patel, A., Vought, V. E., Dharmarajan, V. & Cosgrove, M. S. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 283, 32162–32175 (2008).
Karatas, H. et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein–protein interaction. J. Am. Chem. Soc. 135, 669–682 (2013).
Wilson, C. G. & Arkin, M. R. Small-molecule inhibitors of IL-2/IL-2R: lessons learned and applied. Curr. Top. Microbiol. Immunol. 348, 25–59 (2011).
Tilley, J. W. et al. Identification of a small molecule inhibitor of the IL-2/IL-2Rα receptor interaction which binds to IL-2. J. Am. Chem. Soc. 119, 7589–7590 (1997).
Arkin, M. R. et al. Binding of small molecules to an adaptive protein–protein interface. Proc. Natl Acad. Sci. USA 100, 1603–1608 (2003). In this paper the flexibility and adaptivity of certain protein–protein interfaces is highlighted, with a small molecule that binds a pocket in IL-2 that is not seen in the unbound IL-2 structure. The technique of fragment 'tethering' was then applied to find other hits for this cryptic pocket.
Braisted, A. C. et al. Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. J. Am. Chem. Soc. 125, 3714–3715 (2003). The first description of 'tethering' to discover a high potency inhibitor against a PPI interface.
Raimundo, B. C. et al. Integrating fragment assembly and biophysical methods in the chemical advancement of small-molecule antagonists of IL-2: an approach for inhibiting protein–protein interactions. J. Med. Chem. 47, 3111–3130 (2004).
Hyde, J., Braisted, A. C., Randal, M. & Arkin, M. R. Discovery and characterization of cooperative ligand binding in the adaptive region of interleukin-2. Biochemistry 42, 6475–6483 (2003).
Arrowsmith, C. H., Bountra, C., Fish, P. V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug. Discov. 11, 384–400 (2012).
Furdas, S. D., Carlino, L., Sippl, W. & Jung, M. Inhibition of bromodomain-mediated protein–protein interactions as a novel therapeutic strategy. MedChemComm 3, 123–134 (2012).
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). In this paper the authors report a small molecule with high potency and a remarkable selectivity for a small subset of human bromodomains.
Mirguet, O. et al. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J. Med. Chem. 56, 7501–7515 (2013).
Zimmermann, G. et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 497, 638–642 (2013). The first report of small molecules that disrupt KRAS localization to PDEδ by binding in the prenyl binding pocket of PDEδ.
Ku, B., Liang, C., Jung, J. U. & Oh, B. H. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 21, 627–641 (2011).
Bruncko, M. et al. Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J. Med. Chem. 50, 641–662 (2007).
Robb, R. J., Rusk, C. M. & Neeper, M. P. Structure-function relationships for the interleukin 2 receptor: location of ligand and antibody binding sites on the Tac receptor chain by mutational analysis. Proc. Natl Acad. Sci. USA 85, 5654–5658 (1988).
Sauve, K. et al. Localization in human interleukin 2 of the binding site to the alpha chain (p55) of the interleukin 2 receptor. Proc. Natl Acad. Sci. USA 88, 4636–4640 (1991).
Thanos, C. D., DeLano, W. L. & Wells, J. A. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc. Natl Acad. Sci. USA 103, 15422–15427 (2006).
Acknowledgements
The authors thank T. Blundell and H. Jubb for helpful discussions. J.S., D.E.S. and A.R.B. thank the Wellcome Trust for funding.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
J.S. owns shares in GlaxoSmithKline. C.A. is co-founder and advisor for Astex Pharmaceuticals. A.B. is an employee of Vertex.
Glossary
- Globular protein
-
A protein whose peptide chains are folded to form a broadly spherical shape. This term is often extended to mean a non-membrane-bound protein in which multiple regions of the peptide chain combine to give a defined tertiary structure with one or more binding or active sites.
- Hot spots
-
Regions of a binding surface that contribute a disproportionately large amount to the interaction energy of a pair of proteins or a protein and a ligand.
- Alanine scanning
-
A set of experiments in which amino acids in a protein are sequentially mutated to Ala in order to estimate the contribution of the individual side chains to the binding energy of a protein–protein interaction.
- Discontinuous epitope
-
A binding site at which amino acids from different regions of the protein sequence combine to interact with the partner protein. For example, periodic side chains on one face of a helix or residues from a group of neighbouring chains in a globular protein.
- Intrinsically disordered peptide
-
A peptide lacking a fixed 3D structure when in a monomeric state. Such species may adopt a more ordered structure when interacting with other proteins or ligands.
- Druggability
-
An assessment of the ease with which a drug can be developed to interact with a given protein target. In this case, it has a relatively narrow meaning — namely the ease of generating a small molecule with reasonable affinity.
- Pharmacophore
-
The features of a ligand responsible for its binding to a protein, which are reduced to an abstract representation lacking an underlying framework of bonds or rings
- Apoptosis
-
Programmed cell death controlled by a complex network of interactions including protein–protein interactions.
- Ubiquitylation
-
The attachment of a small protein, ubiquitin, onto another protein — often as a prelude to controlled degradation of the labelled protein.
- ADMET
-
Absorption, distribution, metabolism, excretion and toxicity — key properties of a potential drug that depend, in part, on the physicochemical properties of a molecule.
- PDZ domain
-
A protein domain containing a series of Gly-Leu-Gly-Phe repeating units. Named after the postsynaptic density protein 95 (PSD95), the Drosophilia melanogaster tumour suppressor discs large 1 (Dlg1; also known as Dlg-A) and the tight-junction associated protein zonula occludens 1 (ZO1).
Rights and permissions
About this article
Cite this article
Scott, D., Bayly, A., Abell, C. et al. Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat Rev Drug Discov 15, 533–550 (2016). https://doi.org/10.1038/nrd.2016.29
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2016.29
This article is cited by
-
Design principles for cyclin K molecular glue degraders
Nature Chemical Biology (2024)
-
Small-molecule discovery through DNA-encoded libraries
Nature Reviews Drug Discovery (2023)
-
Development of cyclic peptides that can be administered orally to inhibit disease targets
Nature Chemical Biology (2023)
-
Identification of inhibitors targeting the energy-coupling factor (ECF) transporters
Communications Biology (2023)
-
Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales
NPG Asia Materials (2023)