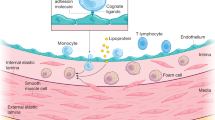
Abstract
Coronary heart disease (CHD) will soon become the leading cause of death and morbidity in the world. Early detection and treatment of CHD is thus imperative to improve global health. Atherosclerosis of the coronary arteries is a complex multifactorial disease process involving multiple pathways that can be influenced by both genetic and environmental factors. With the recent advances in genomics and proteomics, many new risk factors with small-to-moderate effects are likely to be identified. Additionally, individualized risk stratification and targeted therapy may become feasible; each individual could potentially be assessed with a panel of tests for genomic and proteomic markers and, on the basis of the individual's composite risk profile, preventive and therapeutic steps could then be undertaken. With a multimarker approach, it may also be possible to identify alterations in pathways involved in atherogenesis, rather than focus on individual risk factors. In this article, we use the specific example of atherosclerosis to discuss the role of genomics and proteomics in cardiovascular risk assessment.
Key Points
-
Conventional risk factors for atherosclerosis are prevalent in much of the general population and, therefore, available risk-prediction algorithms based on such risk factors have less than desired accuracy
-
New genomic and proteomic markers are likely to have an important role in refining cardiovascular risk assessment, thereby permitting individualized risk stratification and targeted therapy
-
Genome-wide association studies have identified variants that have modest effects on disease propensity; sequencing of entire genomes, or parts thereof, might allow identification of rare variants with stronger effects
-
Proteomic discovery technology is improving and holds promise for identifying new biomarkers that will refine cardiovascular risk stratification
-
In the future, individuals could potentially be assessed with a panel of tests for the many new genomic and proteomic markers that each have small-to-moderate effects
-
A multimarker approach might also enable us to identify alterations in specific pathways of atherogenesis as a means to assess cardiovascular risk, rather than assessing risk by focusing on individual risk factors
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Murray, C. J. L. & Lopez, A. D. Alternative projections of mortality and disability by cause 1990–2020: global burden of disease study. Lancet 349, 1498–1504 (1997).
Yusuf, S., Reddy, S., Ounpuu, S. & Anand, S. Global burden of cardiovascular diseases: Part II: variations in cardiovascular disease by specific ethnic groups and geographic regions and prevention strategies. Circulation 104, 2855–2864 (2001).
Executive Summary of the Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). JAMA 285, 2486–2497 (2001).
Wilson, P. W. et al. Prediction of coronary heart disease using risk factor categories. Circulation 97, 1837–1847 (1998).
Kullo, I. J. & Ballantyne, C. M. Conditional risk factors for atherosclerosis. Mayo Clin. Proc. 80, 219–230 (2005).
Cooper, J. A., Miller, G. J. & Humphries, S. E. A comparison of the PROCAM and Framingham point-scoring systems for estimation of individual risk of coronary heart disease in the Second Northwick Park Heart Study. Atherosclerosis 181, 93–100 (2005).
Wald, N. J., Hackshaw, A. K. & Frost, C. D. When can a risk factor be used as a worthwhile screening test? BMJ 319, 1562–1565 (1999).
Fuster, V., Lois, F. & Franco, M. Early identification of atherosclerotic disease by noninvasive imaging. Nat. Rev. Cardiol. doi:10.1038/nrcardio.2010.54.
Mayr, M. Metabolomics: ready for the prime time? Circ. Cardiovasc. Genet. 1, 58–65 (2008).
Morrison, A. C. et al. Prediction of coronary heart disease risk using a genetic risk score: The Atherosclerosis Risk in Communities Study. Am. J. Epidemiol. 166, 28–35 (2007).
Khoury, M. J., Jones, K. & Grosse, S. D. Quantifying the health benefits of genetic tests: the importance of a population perspective. Genet. Med. 8, 191–195 (2006).
Cortese, D. A. A vision of individualized medicine in the context of global health. Clin. Pharmacol. Ther. 82, 491–493 (2007).
Kim, C. X. et al. Sex and ethnic differences in 47 candidate proteomic markers of cardiovascular disease: The Mayo Clinic Proteomic Markers of Arteriosclerosis Study. PLoS ONE 5, e9065 (2010).
Lusis, A. J. Atherosclerosis. Nature 407, 233–241 (2000).
Guttmacher, A. E., Collins, F. S. & Carmona, R. H. The family history—more important than ever. N. Engl. J. Med. 351, 2333–2336 (2004).
Scheuner, M. T. Family history: where to go from here. Genet. Med. 5, 66–68 (2003).
Kullo, I. J. & Ding, K. Mechanisms of disease: The genetic basis of coronary heart disease. Nat. Clin. Pract. Cardiovasc. Med. 4, 558–569 (2007).
Williams, R. R. et al. Usefulness of cardiovascular family history data for population-based preventive medicine and medical research (the Health Family Tree Study and the NHLBI Family Heart Study). Am. J. Cardiol. 87, 129–135 (2001).
Murabito, J. M. et al. Sibling cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults. JAMA 294, 3117–3123 (2005).
Utermann, G. et al. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature 269, 604–607 (1977).
Clarke, R. et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361, 2518–2528 (2009).
Cohen, J. C., Boerwinkle, E., Mosley, T. H. Jr & Hobbs, H. H. Sequence variations in pcsk9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354, 1264–1272 (2006).
Kullo, I. J. et al. Association of polymorphisms in NOS3 with the ankle-brachial index in hypertensive adults. Atherosclerosis 196, 905–912 (2008).
Ng, S. B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Ding, K. & Kullo, I. J. Genome-wide association studies for atherosclerotic vascular disease and its risk factors. Circ. Cardiovasc. Genet. 2, 63–72 (2009).
Visel, A. et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature 464, 409–412 (2010).
Samani, N. J. et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 357, 443–453 (2007).
Ragoussis, J. Genotyping technologies for genetic research. Annu. Rev. Genomics Hum. Genet. 10, 117–133 (2009).
Pollex, R. L. & Hegele, R. A. Copy number variation in the human genome and its implications for cardiovascular disease. Circulation 115, 3130–3138 (2007).
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
D'Agostino, R. B., Sr et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation 117, 743–753 (2008).
Gail, M. H. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J. Natl Cancer Inst. 100, 1037–1041 (2008).
Janssens, A. C. & van Duijn, C. M. Genome-based prediction of common diseases: advances and prospects. Hum. Mol. Genet. 17, R166–R173 (2008).
Talmud, P. J. et al. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin. Chem. 54, 467–474 (2008).
Brautbar, A. et al. Impact of adding a single allele in the 9p21 locus to traditional risk factors on risk classification for coronary heart disease and implications for lipid-modifying therapy in the white population of the Atherosclerosis Risk in Communities (ARIC) study. Circ. Cardiovasc. Genet. 2, 279–285 (2009).
Paynter, N. P. et al. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann. Intern. Med. 150, 65–72 (2009).
Thomas, D. Gene–environment-wide association studies: emerging approaches. Nat. Rev. Genet. 11, 259–272 (2010).
Hunter, D. J., Khoury, M. J. & Drazen, J. M. Letting the genome out of the bottle—will we get our wish? N. Engl. J. Med. 358, 105–107 (2008).
Khoury, M. J. et al. The Scientific Foundation for personal genomics: recommendations from a National Institutes of Health-Centers for Disease Control and Prevention multidisciplinary workshop. Genet. Med. 11, 559–567 (2009).
McGuire, A. L. & Burke, W. An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA 300, 2669–2671 (2008).
Weston, A. D. & Hood, L. Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J. Proteome Res. 3, 179–196 (2004).
Haddow, J. & Palomaki, G. ACCE: A model process for evaluating data on emerging genetic tests (Oxford University Press, New York, 2004).
Khawaja, F. J. et al. Association of novel risk factors with the ankle brachial index in African American and non-Hispanic white populations. Mayo Clin. Proc. 82, 709–716 (2007).
Pepe, M. S. An interpretation for the ROC curve and inference using GLM procedures. Biometrics 56, 352–359 (2000).
Domon, B. & Aebersold, R. Mass spectrometry and protein analysis. Science 312, 212–217 (2006).
Rifai, N., Gillette, M. A. & Carr, S. A. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 (2006).
Anderson, N. L. et al. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res. 3, 235–244 (2004).
Keshishian, H., Addona, T., Burgess, M., Kuhn, E. & Carr, S. A. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell Proteomics 6, 2212–2229 (2007).
Ling, M. M., Ricks, C. & Lea, P. Multiplexing molecular diagnostics and immunoassays using emerging microarray technologies. Expert Rev. Mol. Diagn. 7, 87–98 (2007).
Schmidt, A. M., Yan, S. D., Wautier, J. L. & Stern, D. Activation of receptor for advanced glycation end products: a mechanism for chronic vascular dysfunction in diabetic vasculopathy and atherosclerosis. Circ. Res. 84, 489–497 (1999).
Cooper, L. T. Jr, et al. Genomic and proteomic analysis of myocarditis and dilated cardiomyopathy. Heart Fail. Clin. 6, 75–85 (2010).
Granger, C. B., Van Eyk, J. E., Mockrin, S. C. & Anderson, N. L. National Heart, Lung, and Blood Institute Clinical Proteomics Working Group Report. Circulation 109, 1697–1703 (2004).
Department of Health and Human Services, Centers for Disease Control and Prevention. Current CLIA Regulations (including all changes through 01/24/2004) [online], (2004).
Ellington, A. A., Kullo, I. J., Bailey, K. R. & Klee, G. G. Antibody-based protein multiplex platforms: technical and operational challenges. Clin. Chem. 56, 186–193 (2009).
Ellington, A. A., Kullo, I. J., Bailey, K. R. & Klee, G. G. Measurement and quality control issues in multiplex protein assays: a case study. Clin. Chem. 55, 1092–1099 (2009).
Kingsmore, S. F. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat. Rev. Drug Discov. 5, 310–320 (2006).
US Food and Drugs Administration. Draft guidance for industry and FDA staff—pharmacogenetic tests and genetic tests for heritable markers [online], (2006).
Rosen, S. in Lateral Flow Immunoassay (eds Wong, R. C. & Tse, H. Y.) 35–49 (Humana Press, New York, 2009).
Wang, T. J. et al. Multiple biomarkers for the prediction of first major cardiovascular events and death. N. Engl. J. Med. 355, 2631–2639 (2006).
Zethelius, B. et al. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N. Engl. J. Med. 358, 2107–2116 (2008).
Cook, N. R. Statistical evaluation of prognostic versus diagnostic models: beyond the ROC curve. Clin. Chem. 54, 17–23 (2008).
Ware, J. H. The limitations of risk factors as prognostic tools. N. Engl. J. Med. 355, 2615–2617 (2006).
Cook, N. R. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation 115, 928–935 (2007).
Pencina, M. J., D'Agostino, R. B., Sr, D'Agostino, R. B. Jr, & Vasan, R. S. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat. Med. 27, 157–172 (2008).
Wang, T. J. New cardiovascular risk factors exist, but are they clinically useful? Eur. Heart J. 29, 441–444 (2008).
Prainsack, B. et al. Personal genomes: Misdirected precaution. Nature 456, 34–35 (2008).
Mosca, L. C-reactive protein—to screen or not to screen? N. Engl. J. Med. 347, 1615–1617 (2002).
Hackam, D. G. & Anand, S. S. Emerging risk factors for atherosclerotic vascular disease: a critical review of the evidence. JAMA 290, 932–940 (2003).
Manolio, T. Novel risk markers and clinical practice. N. Engl. J. Med. 349, 1587–1589 (2003).
Braunwald, E. The Simon Dack lecture. Cardiology: the past, the present, and the future. J. Am. Coll. Cardiol. 42, 2031–2041 (2003).
National Center for Biotechnology Information. The database of Genotypes and Phenotypes (dbGaP) [online], (2010).
Helgadottir, A. et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 (2007).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Coronary Artery Disease Consortium et al. Large scale association analysis of novel genetic loci for coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 29, 774–780 (2009).
Kathiresan, S. et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 (2009).
Willer, C. J. et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169 (2008).
Erdmann, J. et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat. Genet. 41, 280–282 (2009).
Gudbjartsson, D. F. et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 41, 342–347 (2009).
Acknowledgements
This work was supported in part by grants HL81331 and HG04599 from the NIH, USA, and a generous gift from the Marriot family.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kullo, I., Cooper, L. Early identification of cardiovascular risk using genomics and proteomics. Nat Rev Cardiol 7, 309–317 (2010). https://doi.org/10.1038/nrcardio.2010.53
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrcardio.2010.53
This article is cited by
-
LDL-receptor gene polymorphism as a predictor of coronary artery disease: an Egyptian pilot study: relation to lipid profile and angiographic findings
The Egyptian Heart Journal (2024)
-
Systems healthcare: a holistic paradigm for tomorrow
BMC Systems Biology (2017)
-
Motivation, Perception, and Treatment Beliefs in the Myocardial Infarction Genes (MI‐GENES) Randomized Clinical Trial
Journal of Genetic Counseling (2017)
-
Plasma proteomic analysis of stable coronary artery disease indicates impairment of reverse cholesterol pathway
Scientific Reports (2016)
-
Design of a randomized controlled trial of disclosing genomic risk of coronary heart disease: the Myocardial Infarction Genes (MI-GENES) study
BMC Medical Genomics (2015)