Abstract
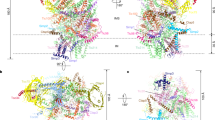
Chloroplasts divide by binary fission, which is accomplished by the simultaneous constriction of the FtsZ ring on the stromal side of the inner envelope membrane, and the ARC5 ring on the cytosolic side of the outer envelope membrane. The two rings are connected and coordinated mainly by the interaction between the inner envelope membrane protein ARC6 and the outer envelope membrane protein PDV2 in the intermembrane space. The underlying mechanism of this coordination is unclear to date. Here, we solved the crystal structure of the intermembrane space region of the ARC6–PDV2 complex. The results indicated that PDV2 inserts its carboxy terminus into a pocket formed in ARC6, and this interaction further induces the dimerization of the intermembrane space regions of two ARC6 molecules. A pdv2 mutant attenuating PDV2-induced ARC6 dimerization showed abnormal morphology of ARC6 rings and compromised chloroplast division in plant cells. Together, our data reveal that PDV2-induced dimerization of ARC6 plays a critical role in chloroplast division and provide insights into the coordination mechanism of the internal and external plastid division machineries.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Osteryoung, K. W. & Pyke, K. A. Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 65, 443–472 (2014).
Yoshida, Y., Miyagishima, S. Y., Kuroiwa, H. & Kuroiwa, T. The plastid-dividing machinery: formation, constriction and fission. Curr. Opin. Plant Biol. 15, 714–721 (2012).
Miyagishima, S. Y., Nakanishi, H. & Kabeya, Y. Structure, regulation, and evolution of the plastid division machinery. Int. Rev. Cell Mol. Biol. 291, 115–153 (2011).
Miyagishima, S. Y. Mechanism of plastid division: from a bacterium to an organelle. Plant Physiol. 155, 1533–1544 (2011).
Yoshida, Y. et al. Isolated chloroplast division machinery can actively constrict after stretching. Science 313, 1435–1438 (2006).
Yoshida, Y. et al. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science 329, 949–953 (2010).
Miyagishima, S. et al. Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13, 2257–2268 (2001).
Vitha, S. et al. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15, 1918–1933 (2003).
Maple, J. & Moller, S. G. Plastid division: evolution, mechanism and complexity. Ann. Bot. 99, 565–579 (2007).
Yang, Y., Glynn, J. M., Olson, B. J., Schmitz, A. J. & Osteryoung, K. W. Plastid division: across time and space. Curr. Opin. Plant Biol. 11, 577–584 (2008).
Osteryoung, K. W. & Pyke, K. A. Plastid division: evidence for a prokaryotically derived mechanism. Curr. Opin. Plant Biol. 1, 475–479 (1998).
Stokes, K. D., McAndrew, R. S., Figueroa, R., Vitha, S. & Osteryoung, K. W. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124, 1668–1677 (2000).
Gao, H., Kadirjan-Kalbach, D., Froehlich, J. E. & Osteryoung, K. W. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl Acad. Sci. USA 100, 4328–4333 (2003).
Glynn, J. M., Froehlich, J. E. & Osteryoung, K. W. Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20, 2460–2470 (2008).
Miyagishima, S. Y., Froehlich, J. E. & Osteryoung, K. W. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18, 2517–2530 (2006).
Maple, J., Aldridge, C. & Moller, S. G. Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 43, 811–823 (2005).
Sugase, K., Dyson, H. J. & Wright, P. E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 (2007).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Kumar, N., Radhakrishnan, A., Su, C. C., Osteryoung, K. W. & Yu, E. W. Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6. Protein Sci. 25, 523–529 (2016).
Handa, N. et al. Structure of the human Tim44 C-terminal domain in complex with pentaethylene glycol: ligand-bound form. Acta Crystallogr. D Biol. Crystallogr. 63, 1225–1234 (2007).
Bullock, T. L., Clarkson, W. D., Kent, H. M. & Stewart, M. The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2). J. Mol. Biol. 260, 422–431 (1996).
Rellos, P. et al. Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol. 8, e1000426 (2010).
Guo, F. et al. Structural insights into the tumor-promoting function of the MTDH-SND1 complex. Cell Rep. 8, 1704–1713 (2014).
Zhang, Q., Jia, N., Li, H. & Gao, H.-B. Identification and analysis of ARC6 gene in a chloroplast division mutant cpd44 in Arabidopsis thaliana. Plant Physiol. J. 51, 8 (2015).
Stewart, M., Kent, H. M. & McCoy, A. J. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J. Mol. Biol. 277, 635–646 (1998).
Guo, F. et al. Structural basis of PP2A activation by PTPA, an ATP-dependent activation chaperone. Cell Res. 24, 190–203 (2014).
Du, D. et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509, 512–515 (2014).
Hu, M. et al. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 4, e27 (2006).
Saredi, G. et al. H4k20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex. Nature 534, 714–718 (2016).
Zhang, M., Chen, C., Froehlich, J. E., TerBush, A. D. & Osteryoung, K. W. Roles of Arabidopsis PARC6 in coordination of the chloroplast division complex and negative regulation of FtsZ assembly. Plant Physiol. 170, 250–262 (2016).
Youle, R. J. & van der Bliek, A. M. Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012).
Marbouty, M., Saguez, C., Cassier-Chauvat, C. & Chauvat, F. Zipn, an FtsA-like orchestrator of divisome assembly in the model cyanobacterium synechocystis PCC6803. Mol. Microbiol. 74, 409–420 (2009).
Shiomi, D. & Margolin, W. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol. Microbiol. 66, 1396–1415 (2007).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. Pt A, 307–326 (1997).
CP Collaborative. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Cowtan, K. D. & Zhang, K. Y. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 72, 245–270 (1999).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Gao, Y. et al. Arabidopsis FRS4/CPD25 and FHY3/CPD45 work cooperatively to promote the expression of the chloroplast division gene ARC5 and chloroplast division. Plant J. 75, 795–807 (2013).
Lukowitz, W., Gillmor, C. S. & Scheible, W. R. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–805 (2000).
Li, Y., Sun, Q., Feng, Y., Liu, X. & Gao, H. An improved immunofluorescence staining method for chloroplast proteins. Plant Cell Rep. 35, 2285–2293 (2016).
Acknowledgements
We would like to thank the staff at beamline BL17U of the Shanghai Synchrotron Radiation Facility for their assistance with data collection, and J. Wang at Tsinghua University for crystallographic data analysis. This work was supported by the National Natural Science Foundation of China (31400635 and 30971439), the Beijing Natural Science Foundation (5154031) and the Fundamental Research Funds for the Central Universities (JD1609).
Author information
Authors and Affiliations
Contributions
Y.F. and H.G. designed the project. W.W., X.Y., W.Z., Y.D., F.H., Y.Y., S.F., M.Y., N.C., Z.Z. and S.L. purified the proteins, grew the crystals, collected data, solved the crystal structures and performed the in vitro pull-down and gel filtration assays. J.L., N.J., X.L. and N.C. performed the yeast two-hybrid assays. J.L., Q.S., C.A., X.L. and N.C. identified the Arabidopsis mutants, generated transgenic plants and did the microscopy and other analyses. Q.S. and Y.L. performed the immunofluorescence staining experiments. Y.F. analysed the data and wrote the paper with the help of all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Figures 1–10, Supplementary Table 1. (PDF 1790 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Li, J., Sun, Q. et al. Structural insights into the coordination of plastid division by the ARC6–PDV2 complex. Nature Plants 3, 17011 (2017). https://doi.org/10.1038/nplants.2017.11
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2017.11
This article is cited by
-
Mutation of CsARC6 affects fruit color and increases fruit nutrition in cucumber
Theoretical and Applied Genetics (2023)
-
Electron Tomography Analysis of Thylakoid Assembly and Fission in Chloroplasts of a Single-Cell C4 plant, Bienertia sinuspersici
Scientific Reports (2019)
-
Chloroplast division: A handshake across membranes
Nature Plants (2017)
-
The beauty of the small
Nature Plants (2017)