Abstract
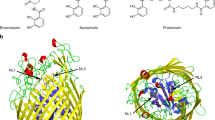
Pathogenic microorganisms must cope with extremely low free-iron concentrations in the host's tissues. Some fungal pathogens rely on secreted haemophores that belong to the Common in Fungal Extracellular Membrane (CFEM) protein family, to extract haem from haemoglobin and to transfer it to the cell's interior, where it can serve as a source of iron. Here we report the first three-dimensional structure of a CFEM protein, the haemophore Csa2 secreted by Candida albicans. The CFEM domain adopts a novel helical-basket fold that consists of six α-helices, and is uniquely stabilized by four disulfide bonds formed by its eight signature cysteines. The planar haem molecule is bound between a flat hydrophobic platform located on top of the helical basket and a peripheral N-terminal ‘handle’ extension. Exceptionally, an aspartic residue serves as the CFEM axial ligand, and so confers coordination of Fe3+ haem, but not of Fe2+ haem. Histidine substitution mutants of this conserved Asp acquired Fe2+ haem binding and retained the capacity to extract haem from haemoglobin. However, His-substituted CFEM proteins were not functional in vivo and showed disturbed haem exchange in vitro, which suggests a role for the oxidation-state-specific Asp coordination in haem acquisition by CFEM proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Weinberg, E. D. Nutritional immunity. Host's attempt to withhold iron from microbial invaders. J. Am. Med. Assoc. 231, 39–41 (1975).
Andrews, N. C. Iron homeostasis: insights from genetics and animal models. Nat. Rev. Genet. 1, 208–217 (2000).
Almeida, R. S., Wilson, D. & Hube, B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 9, 1000–1012 (2009).
Cassat, J. E. & Skaar, E. P. Iron in infection and immunity. Cell Host Microbe 13, 509–519 (2013).
Caza, M. & Kronstad, J. W. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell Infect. Microbiol. 3, 80 (2013).
Letoffe, S., Ghigo, J. M. & Wandersman, C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc. Natl Acad. Sci. USA 91, 9876–9880 (1994).
Ekworomadu, M. T. et al. Differential function of lip residues in the mechanism and biology of an anthrax hemophore. PLoS Pathog. 8, e1002559 (2012).
Cadieux, B. et al. The mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J. Infect. Dis. 207, 1339–1347 (2013).
Kulkarni, R. D., Kelkar, H. S. & Dean, R. A. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28, 118–121 (2003).
Mitchell, A. et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221 (2015).
Ding, C. et al. Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS One 6, e28151 (2011).
Kuznets, G. et al. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog. 10, e1004407 (2014).
Weissman, Z. & Kornitzer, D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 53, 1209–1220 (2004).
Bailao, E. F. et al. Hemoglobin uptake by Paracoccidioides spp. is receptor-mediated. PLoS Negl. Trop. Dis. 8, e2856 (2014).
Amorim-Vaz, S. et al. RNA enrichment method for quantitative transcriptional analysis of pathogens in vivo applied to the fungus Candida albicans. mBio 6, e00942–15 (2015).
Mochon, A. B. et al. Serological profiling of a Candida albicans protein microarray reveals permanent host–pathogen interplay and stage-specific responses during candidemia. PLoS Pathog. 6, e1000827 (2010).
Sorgo, A. G. et al. Mass spectrometric analysis of the secretome of Candida albicans. Yeast 27, 661–672 (2010).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Li, T., Bonkovsky, H. L. & Guo, J. T. Structural analysis of heme proteins: implications for design and prediction. BMC Struct. Biol. 11, 13 (2011).
Outten, F. W. & Theil, E. C. Iron-based redox switches in biology. Antioxid. Redox Signal 11, 1029–1046 (2009).
Dixon, H. B. & McIntosh, R. Reduction of methaemoglobin in haemoglobin samples using gel filtration for continuous removal of reaction products. Nature 213, 399–400 (1967).
Contreras, H., Chim, N., Credali, A. & Goulding, C. W. Heme uptake in bacterial pathogens. Curr. Opin. Chem. Biol. 19, 34–41 (2014).
Arnoux, P. et al. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat. Struct. Biol. 6, 516–520 (1999).
Mazmanian, S. K. et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909 (2003).
Grigg, J. C., Ukpabi, G., Gaudin, C. F. & Murphy, M. E. Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Inorg. Biochem. 104, 341–348 (2010).
Grigg, J. C., Vermeiren, C. L., Heinrichs, D. E. & Murphy, M. E. Haem recognition by a Staphylococcus aureus NEAT domain. Mol. Microbiol. 63, 139–149 (2007).
Bodanszky, M., Klausner, Y. S. & Said, S. I. Biological activities of synthetic peptides corresponding to fragments of and to the entire sequence of the vasoactive intestinal peptide. Proc. Natl Acad. Sci. USA 70, 382–384 (1973).
Weissman, Z., Shemer, R. & Kornitzer, D. Deletion of the copper transporter CaCCC2 reveals two distinct pathways for iron acquisition in Candida albicans. Mol. Microbiol. 44, 1551–1560 (2002).
Atir-Lande, A., Gildor, T. & Kornitzer, D. Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol. Biol. Cell 16, 2772–2785 (2005).
Feng, Q., Summers, E., Guo, B. & Fink, G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181, 6339–6346 (1999).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000 the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006).
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002).
Otwinowski, Z. in Proc. CCP4 Study Weekend. Isomorphous Replacement and Anomalous Scattering (eds Wolf, W., Evans, P. R. & Leslie, A. G. W. ) 80–85 (Daresbury Laboratory, 1991).
Cowtan, K. D. & Zhang, K. Y. Density modification for macromolecular phase improvement. Progr. Biophys. Mol. Biol. 72, 245–270 (1999).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Weinstein, J. D. & Beale, S. I. Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis. J. Biol. Chem. 258, 6799–6807 (1983).
Schneider, S., Marles-Wright, J., Sharp, K. H. & Paoli, M. Diversity and conservation of interactions for binding heme in b-type heme proteins. Nat. Prod. Rep. 24, 621–630 (2007).
Zacharias, N. & Dougherty, D. A. Cation–pi interactions in ligand recognition and catalysis. Trends Pharmacol. Sci. 23, 281–287 (2002).
Hargrove, M. S., Whitaker, T., Olson, J. S., Vali, R. J. & Mathews, A. J. Quaternary structure regulates hemin dissociation from human hemoglobin. J. Biol. Chem. 272, 17385–9 (1997).
Acknowledgements
We are grateful to D. Hiya (Technion Center for Structural Biology) for help with protein crystallization, T. Ziv (Smoler Proteomics Center) for mass spectroscopy analysis, G. Kuznets and Y. Gorelik for constructing plasmids KB2366 and KB2221/2, U. Roy for producing Csa2 cysteine mutants, M. Lebendiker (Hebrew University of Jerusalem) for advice with SEC-MALS, R. Zarivach (Ben-Gurion University) for the Origami B strain and O. Lewinson, I. Silman, N. Adir, N. Levanon, A. Haber and S. Selig for discussions and critical reading of the manuscript. This research was supported by grants from the Israel Science Foundation and the Ministry of Health's Chief Scientist Office to D.K. H.D. thanks the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement No. 330879-MC-CHOLESTRUCTURE for financial support.
Author information
Authors and Affiliations
Contributions
L.N., Z.W., M.P. and D.K. constructed the plasmids and strains, purified the proteins and performed experiments, H.A. performed the SEC-MALS analysis, H.D. crystallized the protein and determined its structure, and H.D. and D.K. developed the project, interpreted the data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–9, Supplementary Tables 1–3, Supplementary References (PDF 4321 kb)
Rights and permissions
About this article
Cite this article
Nasser, L., Weissman, Z., Pinsky, M. et al. Structural basis of haem-iron acquisition by fungal pathogens. Nat Microbiol 1, 16156 (2016). https://doi.org/10.1038/nmicrobiol.2016.156
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.156
This article is cited by
-
Template-Based Modelling of the Structure of Fungal Effector Proteins
Molecular Biotechnology (2023)
-
Comparative roles of three adhesin genes (adh1–3) in insect-pathogenic lifecycle of Beauveria bassiana
Applied Microbiology and Biotechnology (2021)
-
Machinery for fungal heme acquisition
Current Genetics (2020)
-
Overview of Alternaria alternata Membrane Proteins
Indian Journal of Microbiology (2020)
-
In vivo gene expression profiling of the entomopathogenic fungus Beauveria bassiana elucidates its infection stratagems in Anopheles mosquito
Science China Life Sciences (2017)