Abstract
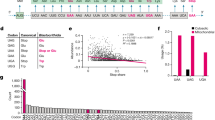
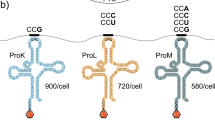
Although regulation of translation fidelity is an essential process1–7, diverse organisms and organelles have differing requirements of translational accuracy8–15, and errors in gene translation serve an adaptive function under certain conditions16–20. Therefore, optimal levels of fidelity may vary according to context. Most bacteria utilize a two-step pathway for the specific synthesis of aminoacylated glutamine and/or asparagine tRNAs, involving the glutamine amidotransferase GatCAB21–25, but it had not been appreciated that GatCAB may play a role in modulating mistranslation rates. Here, by using a forward genetic screen, we show that the mycobacterial GatCAB enzyme complex mediates the translational fidelity of glutamine and asparagine codons. We identify mutations in gatA that cause partial loss of function in the holoenzyme, with a consequent increase in rates of mistranslation. By monitoring single-cell transcription dynamics, we demonstrate that reduced gatCAB expression leads to increased mistranslation rates, which result in enhanced rifampicin-specific phenotypic resistance. Consistent with this, strains with mutations in gatA from clinical isolates of Mycobacterium tuberculosis show increased mistranslation, with associated antibiotic tolerance, suggesting a role for mistranslation as an adaptive strategy in tuberculosis. Together, our findings demonstrate a potential role for the indirect tRNA aminoacylation pathway in regulating translational fidelity and adaptive mistranslation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bullwinkle, T. J. et al. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. Elife 3, e02501 (2014).
LaRiviere, F. J., Wolfson, A. D. & Uhlenbeck, O. C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294, 165–168 (2001).
Lee, J. W. et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55 (2006).
Liu, Y. et al. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc. Natl Acad. Sci. USA 111, 17570–17575 (2014).
Lu, J., Bergert, M., Walther, A. & Suter, B. Double-sieving-defective aminoacyl-tRNA synthetase causes protein mistranslation and affects cellular physiology and development. Nat. Commun. 5, 5650 (2014).
Reynolds, N. M., Lazazzera, B. A. & Ibba, M. Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 8, 849–856 (2010).
Zaher, H. S. & Green, R. Quality control by the ribosome following peptide bond formation. Nature 457, 161–166 (2009).
Leng, T., Pan, M., Xu, X. & Javid, B. Translational misreading in Mycobacterium smegmatis increases in stationary phase. Tuberculosis 95, 678–681 (2015).
Li, L. et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl Acad. Sci. USA 108, 9378–9383 (2011).
Ling, J., O'Donoghue, P. & Soll, D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat. Rev. Microbiol. 13, 707–721 (2015).
Netzer, N. et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526 (2009).
Reynolds, N. M. et al. Cell-specific differences in the requirements for translation quality control. Proc. Natl Acad. Sci. USA 107, 4063–4068 (2010).
Ribas de Pouplana, L., Santos, M. A., Zhu, J. H., Farabaugh, P. J. & Javid, B. Protein mistranslation: friend or foe? Trends Biochem. Sci. 39, 355–362 (2014).
Ruan, B. et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl Acad. Sci. USA 105, 16502–16507 (2008).
Yadavalli, S. S. & Ibba, M. Selection of tRNA charging quality control mechanisms that increase mistranslation of the genetic code. Nucleic Acids Res. 41, 1104–1112 (2013).
Bezerra, A. R. et al. Reversion of a fungal genetic code alteration links proteome instability with genomic and phenotypic diversification. Proc. Natl Acad. Sci. USA 110, 11079–11084 (2013).
Fan, Y. et al. Protein mistranslation protects bacteria against oxidative stress. Nucleic Acids Res. 43, 1740–1748 (2015).
Javid, B. et al. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc. Natl Acad. Sci. USA 111, 1132–1137 (2014).
Lee, J. Y. et al. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J. Cell Sci. 127, 4234–4245 (2014).
Miranda, I. et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio 4, e00285-13 (2013).
Bailly, M., Blaise, M., Lorber, B., Becker, H. D. & Kern, D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell 28, 228–239 (2007).
Curnow, A. W. et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA 94, 11819–11826 (1997).
Huot, J. L. et al. Gln-tRNAGln synthesis in a dynamic transamidosome from Helicobacter pylori, where GluRS2 hydrolyzes excess Glu-tRNAGln. Nucleic Acids Res. 39, 9306–9315 (2011).
Nakamura, A., Yao, M., Chimnaronk, S., Sakai, N. & Tanaka, I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science 312, 1954–1958 (2006).
Silva, G. N. et al. A tRNA-independent mechanism for transamidosome assembly promotes aminoacyl-tRNA transamidation. J. Biol. Chem. 288, 3816–3822 (2013).
Manickam, N., Nag, N., Abbasi, A., Patel, K. & Farabaugh, P. J. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA 20, 9–15 (2014).
Wong, S. Y. et al. Functional role of methylation of G518 of the 16S rRNA 530 loop by GidB in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 6311–6318 (2013).
Roy, H., Becker, H. D., Mazauric, M. H. & Kern, D. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 35, 3420–3430 (2007).
Adams, K. N. et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53 (2011).
Aldridge, B. B. et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335, 100–104 (2012).
Comas, I. et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45, 1176–1182 (2013).
Dhar, N. & McKinney, J. D. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 10, 30–38 (2007).
Fridman, O., Goldberg, A., Ronin, I., Shoresh, N. & Balaban, N. Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
Maisonneuve, E. & Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 (2014).
Shah, D. et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6, 53 (2006).
Shan, Y., Lazinski, D., Rowe, S., Camilli, A. & Lewis, K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. MBio 6, e00078-15 (2015).
Siatecka, M., Rozek, M., Barciszewski, J. & Mirande, M. Modular evolution of the Glx-tRNA synthetase family—rooting of the evolutionary tree between the bacteria and archaea/eukarya branches. Eur. J. Biochem. 256, 80–87 (1998).
Campbell, E. A. et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104, 901–912 (2001).
van Kessel, J. C. & Hatfull, G. F. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol. Microbiol. 67, 1094–1107 (2008).
Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T. & Jacobs, W. R. Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919 (1990).
Siegrist, M. S. et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl Acad. Sci. USA 106, 18792–18797 (2009).
Parish, T. & Stoker, N. G. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975 (2000).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Gagneux, S. et al. Variable host–pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 2869–2873 (2006).
Koser, C. U. et al. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. New Engl. J. Med. 369, 290–292 (2013).
Acknowledgements
This work was funded in part by the Bill and Melinda Gates Foundation (OPP1109789) and start-up funds from Tsinghua University to B.J. B.J. and T.F.Z. are Tsinghua-Janssen scholars. The authors thank S. Fortune, E. Rubin, M. Chao and P. Lehner for their critical reading of the manuscript, and C. Koser and P. Abel Zur Wiesch for helpful discussions. The authors also acknowledge technical assistance from the microbial sorting facility at Peking University, and thank the Clinical Database and Sample Bank of Tuberculosis of Beijing (D131100005313012) of the National Clinical Lab on Tuberculosis, Beijing Chest Hospital, for access to their strain collection.
Author information
Authors and Affiliations
Contributions
B.J. conceived and oversaw the design and implementation of the project. H.W.S., J.H.Z., C.E., X.W. and H.L. designed and performed the research and analysed the data. R.J.C. and Y.X.C. made and provided reagents. M.K., T.F.Z. and D.M. analysed whole-genome sequencing data. H.H., B.D.K. and B.J. analysed data. H.W.S., J.H.Z. and B.J. wrote the paper with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The editors note that one of the individuals acknowledged for critical reading of the manuscript, M.C., as well as being a former colleague of the corresponding author, is an editor on the staff of Nature Microbiology, but was not in any way involved in the journal review process.
Supplementary information
Supplementary information
Supplementary Figures 1–11, Supplementary Tables 1–5, Supplementary References. (PDF 14754 kb)
Supplementary Tables 6–8
List of primers, strains and plasmids used in this study. (XLSX 17 kb)
Rights and permissions
About this article
Cite this article
Su, HW., Zhu, JH., Li, H. et al. The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat Microbiol 1, 16147 (2016). https://doi.org/10.1038/nmicrobiol.2016.147
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.147
This article is cited by
-
Rifamycin antibiotics and the mechanisms of their failure
The Journal of Antibiotics (2021)
-
Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis
Nature Communications (2021)
-
Clinically relevant mutations in mycobacterial LepA cause rifampicin-specific phenotypic resistance
Scientific Reports (2020)
-
HspX promotes the polar localization of mycobacterial protein aggregates
Scientific Reports (2019)
-
More than merely drug resistance
Nature Microbiology (2018)