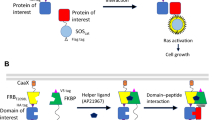
Abstract
Interactions between proteins are at the heart of the cellular machinery. It is therefore not surprising that altered interaction profiles caused by aberrant protein expression patterns or by the presence of mutations can trigger cellular dysfunction, eventually leading to disease. Moreover, many viral and bacterial pathogens rely on protein-protein interactions to exert their damaging effects. Interfering with such interactions is an obvious pharmaceutical goal, but detailed insights into the protein binding properties as well as efficient screening platforms are needed. In this report, we describe a cytokine receptor–based assay with a positive readout to screen for disrupters of designated protein-protein interactions in intact mammalian cells and evaluate this concept using polypeptides as well as small organic molecules. These reverse mammalian protein-protein interaction trap (MAPPIT) screens were developed to monitor interactions between the erythropoietin receptor (EpoR) and suppressors of cytokine signaling (SOCS) proteins, between FKBP12 and ALK4, and between MDM2 and p53.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Zhang, Z.Y., Poorman, R.A., Maggiora, L.L., Heinrikson, R.L. & Kezdy, F.J. Dissociative inhibition of dimeric enzymes. Kinetic characterization of the inhibition of HIV-1 protease by its COOH-terminal tetrapeptide. J. Biol. Chem. 266, 15591–15594 (1991).
Moss, N. et al. Peptidomimetic inhibitors of herpes simplex virus ribonucleotide reductase with improved in vivo antiviral activity. J. Med. Chem. 39, 4173–4180 (1996).
Adachi, T., Stafford, S., Sur, S. & Alam, R. A novel Lyn-binding peptide inhibitor blocks eosinophil differentiation, survival, and airway eosinophilic inflammation. J. Immunol. 163, 939–946 (1999).
Degterev, A. et al. Identification of small-molecule inhibitors of interaction between the BH3 domain and Bcl-xL . Nat. Cell Biol. 3, 173–182 (2001).
Hayashi, M. et al. Suppression of bone resorption by madindoline A, a novel nonpeptide antagonist to gp130. Proc. Natl. Acad. Sci. USA 99, 14728–14733 (2002).
Hayashi, M. et al. Biological activity of a novel nonpeptide antagonist to the interleukin-6 receptor 20S,21-epoxy-resibufogenin-3-formate. J. Pharmacol. Exp. Ther. 303, 104–109 (2002).
Weitz-Schmidt, G. et al. Statins selectively inhibit leukocyte function antigen-1 by binding to a novel regulatory integrin site. Nat. Med. 7, 687–692 (2001).
Vassilev, L.T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Lepourcelet, M. et al. Small-molecule antagonists of the oncogenic Tcf/β-catenin protein complex. Cancer Cell 5, 91–102 (2004).
Gadek, T.R. & Nicholas, J.B. Small molecule antagonists of proteins. Biochem. Pharmacol. 65, 1–8 (2003).
Bergendahl, V., Heyduk, T. & Burgess, R.R. Luminescence resonance energy transfer-based high-throughput screening assay for inhibitors of essential protein-protein interactions in bacterial RNA polymerase. Appl. Environ. Microbiol. 69, 1492–1498 (2003).
Zhao, H.F. et al. A mammalian genetic system to screen for small molecules capable of disrupting protein-protein interactions. Anal. Chem. 76, 2922–2927 (2004).
Kato-Stankiewicz, J. et al. Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. Proc. Natl. Acad. Sci. USA 99, 14398–14403 (2002).
Huang, J. & Schreiber, S.L. A yeast genetic system for selecting small molecule inhibitors of protein-protein interactions in nanodroplets. Proc. Natl. Acad. Sci. USA 94, 13396–13401 (1997).
Vidal, M., Braun, P., Chen, E., Boeke, J.D. & Harlow, E. Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc. Natl. Acad. Sci. USA 93, 10321–10326 (1996).
Park, S.H. & Raines, R.T. Genetic selection for dissociative inhibitors of designated protein-protein interactions. Nat. Biotechnol. 18, 847–851 (2000).
Eyckerman, S. et al. Design and application of a cytokine receptor–based interaction trap. Nat. Cell Biol. 3, 1114–1119 (2001).
Krebs, D.L. & Hilton, D.J. SOCS proteins: negative regulators of cytokine signaling. Stem Cells 19, 378–387 (2001).
Yoshimura, A. et al. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 14, 2816–2826 (1995).
Baumann, H. et al. The full-length leptin receptor has signaling capabilities of interleukin 6-type cytokine receptors. Proc. Natl. Acad. Sci. USA 93, 8374–8378 (1996).
Eyckerman, S., Broekaert, D., Verhee, A., Vandekerckhove, J. & Tavernier, J. Identification of the Y985 and Y1077 motifs as SOCS3 recruitment sites in the murine leptin receptor. FEBS Lett. 486, 33–37 (2000).
Zhang, J.G. et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. USA 96, 2071–2076 (1999).
Bjorbaek, C., Elmquist, J.K., Frantz, J.D., Shoelson, S.E. & Flier, J.S. Identification of SOCS-3 as a potential mediator of central leptin resistance. Mol. Cell 1, 619–625 (1998).
Huse, M., Chen, Y.G., Massague, J. & Kuriyan, J. Crystal structure of the cytoplasmic domain of the type I TGF β receptor in complex with FKBP12. Cell 96, 425–436 (1999).
Wang, T., Donahoe, P.K. & Zervos, A.S. Specific interaction of type I receptors of the TGF-beta family with the immunophilin FKBP-12. Science 265, 674–676 (1994).
Cook, W.S. & Unger, R.H. Protein tyrosine phosphatase 1B: a potential leptin resistance factor of obesity. Dev. Cell 2, 385–387 (2002).
ten Hoeve, J. et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 22, 5662–5668 (2002).
Chung, C.D. et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278, 1803–1805 (1997).
Chen, Y.G., Liu, F. & Massague, J. Mechanism of TGFbeta receptor inhibition by FKBP12. EMBO J. 16, 3866–3876 (1997).
Charng, M.J., Kinnunen, P., Hawker, J., Brand, T. & Schneider, M.D. FKBP-12 recognition is dispensable for signal generation by type I transforming growth factor-βreceptors. J. Biol. Chem. 271, 22941–22944 (1996).
Bogan, A.A. & Thorn, K.S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998).
Archakov, A.I. et al. Protein-protein interactions as a target for drugs in proteomics. Proteomics 3, 380–391 (2003).
Arkin, M.R. et al. Binding of small molecules to an adaptive protein-protein interface. Proc. Natl. Acad. Sci. USA 100, 1603–1608 (2003).
Vidal, M. & Endoh, H. Prospects for drug screening using the reverse two-hybrid system. Trends Biotechnol. 17, 374–381 (1999).
Gaber, R.F., Copple, D.M., Kennedy, B.K., Vidal, M. & Bard, M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 9, 3447–3456 (1989).
Eyckerman, S. et al. Analysis of Tyr to Phe and fa/fa leptin receptor mutations in the PC12 cell line. Eur. Cytokine Netw. 10, 549–556 (1999).
Lemmens, I. et al. Heteromeric MAPPIT: a novel strategy to study modification-dependent protein-protein interactions in mammalian cells. Nucleic Acids Res. 31, e75 (2003).
Acknowledgements
We wish to acknowledge R. Devos for the pGEX-PTP-1B vector; K. Kas for critical reading of the manuscript, M. Goethals for peptide synthesis and D. Defeau for technical assistance. This project was supported by grants from the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT-Vlaanderen; GBOU 010090), Ghent University (UGent; GOA 12051401) and the valorisation fund of the Flanders Interuniversity Institute for Biotechnology (VIB). S.E. is a Postdoctoral Fellow of the Fund for Scientific Research-Flanders (FWO-V).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Dose-dependent disruption of the p53/MDM2 interaction: comparison of different configurations. (PDF 975 kb)
Rights and permissions
About this article
Cite this article
Eyckerman, S., Lemmens, I., Catteeuw, D. et al. Reverse MAPPIT: screening for protein-protein interaction modifiers in mammalian cells. Nat Methods 2, 427–433 (2005). https://doi.org/10.1038/nmeth760
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth760
This article is cited by
-
Trapping mammalian protein complexes in viral particles
Nature Communications (2016)
-
Cheiradone: a vascular endothelial cell growth factor receptor antagonist
BMC Cell Biology (2008)
-
Reverse MAPPIT detects disruptors of protein-protein interactions in human cells
Nature Protocols (2006)
-
Single protein complex visualization: Seeing is believing
Nature Methods (2006)
-
Perturbing interactions
Nature Methods (2005)