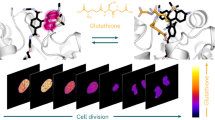
Abstract
GTP is a major regulator of multiple cellular processes, but tools for quantitative evaluation of GTP levels in live cells have not been available. We report the development and characterization of genetically encoded GTP sensors, which we constructed by inserting a circularly permuted yellow fluorescent protein (cpYFP) into a region of the bacterial G protein FeoB that undergoes a GTP-driven conformational change. GTP binding to these sensors results in a ratiometric change in their fluorescence, thereby providing an internally normalized response to changes in GTP levels while minimally perturbing those levels. Mutations introduced into FeoB to alter its affinity for GTP created a series of sensors with a wide dynamic range. Critically, in mammalian cells the sensors showed consistent changes in ratiometric signal upon depletion or restoration of GTP pools. We show that these GTP evaluators (GEVALs) are suitable for detection of spatiotemporal changes in GTP levels in living cells and for high-throughput screening of molecules that modulate GTP levels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Shoji, S., Walker, S.E. & Fredrick, K. Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem. Biol. 4, 93–107 (2009).
Simon, M.I., Strathmann, M.P. & Gautam, N. Diversity of G proteins in signal transduction. Science 252, 802–808 (1991).
Neves, S.R., Ram, P.T. & Iyengar, R. G protein pathways. Science 296, 1636–1639 (2002).
MacMicking, J.D. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25, 601–609 (2004).
Kresse, A. et al. Analyses of murine GBP homology clusters based on in silico, in vitro and in vivo studies. BMC Genomics 9, 158 (2008).
Vestal, D.J. & Jeyaratnam, J.A. The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J. Interferon Cytokine Res. 31, 89–97 (2011).
Downward, J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 (2003).
Guilfoyle, A. et al. Structural basis of GDP release and gating in G protein coupled Fe2+ transport. EMBO J. 28, 2677–2685 (2009).
Marlovits, T.C., Haase, W., Herrmann, C., Aller, S.G. & Unger, V.M. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. USA 99, 16243–16248 (2002).
Belousov, V.V. et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 3, 281–286 (2006).
Berg, J., Hung, Y.P. & Yellen, G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat. Methods 6, 161–166 (2009).
Imamura, H. et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA 106, 15651–15656 (2009).
Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 (1994).
Matsushika, A., Nagashima, A., Goshima, T. & Hoshino, T. Fermentation of xylose causes inefficient metabolic state due to carbon/energy starvation and reduced glycolytic flux in recombinant industrial Saccharomyces cerevisiae. PLoS One 8, e69005 (2013).
Romani, A.M. Cellular magnesium homeostasis. Arch. Biochem. Biophys. 512, 1–23 (2011).
Palmer, A.E., Qin, Y., Park, J.G. & McCombs, J.E. Design and application of genetically encoded biosensors. Trends Biotechnol. 29, 144–152 (2011).
Tantama, M., Hung, Y.P. & Yellen, G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J. Am. Chem. Soc. 133, 10034–10037 (2011).
Turka, L.A., Dayton, J., Sinclair, G., Thompson, C.B. & Mitchell, B.S. Guanine ribonucleotide depletion inhibits T cell activation. Mechanism of action of the immunosuppressive drug mizoribine. J. Clin. Invest. 87, 940–948 (1991).
Wawrzyniak, J.A. et al. A purine nucleotide biosynthesis enzyme guanosine monophosphate reductase is a suppressor of melanoma invasion. Cell Rep. 5, 493–507 (2013).
Gu, J.J. et al. Induction of apoptosis in IL-3-dependent hematopoietic cell lines by guanine nucleotide depletion. Blood 101, 4958–4965 (2003).
Laliberté, J., Yee, A., Xiong, Y. & Mitchell, B.S. Effects of guanine nucleotide depletion on cell cycle progression in human T lymphocytes. Blood 91, 2896–2904 (1998).
Mannava, S. et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle 7, 2392–2400 (2008).
Buckstein, M.H., He, J. & Rubin, H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726 (2008).
Chen, P. et al. A LC-MS/MS method for the analysis of intracellular nucleoside triphosphate levels. Pharm. Res. 26, 1504–1515 (2009).
Qiu, Y. et al. Mycophenolic acid-induced GTP depletion also affects ATP and pyrimidine synthesis in mitogen-stimulated primary human T-lymphocytes. Transplantation 69, 890–897 (2000).
Wang, Y., Tang, L., Li, Z., Lin, Y. & Li, J. In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet-based sensing platform in living cells. Nat. Protoc. 9, 1944–1955 (2014).
Sumita, K. et al. The lipid kinase PI5P4Kβ is an intracellular GTP sensor for metabolism and tumorigenesis. Mol. Cell 61, 187–198 (2016).
Takeuchi, K. et al. Structural reverse genetics study of the PI5P4Kβ-nucleotide complexes reveals the presence of the GTP bioenergetic system in mammalian cells. FEBS J. 283, 3556–3562 (2016).
Mannava, S. et al. Depletion of deoxyribonucleotide pools is an endogenous source of DNA damage in cells undergoing oncogene-induced senescence. Am. J. Pathol. 182, 142–151 (2013).
Bianchi-Smiraglia, A. et al. Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools. Oncogene 36, 84–96 (2016).
Huh, K.H. et al. The role of thioredoxin 1 in the mycophenolic acid-induced apoptosis of insulin-producing cells. Cell Death Dis. 4, e721 (2013).
Malekinejad, H., Moradi, M. & Fink-Gremmels, J. Cytochrome C and caspase-3/7 are involved in mycophenolic acid-induced apoptosis in genetically engineered PC12 neuronal cells expressing the p53 gene. Iran. J. Pharm. Res. 13, 191–198 (2014).
Zhang, J.H., Chung, T.D. & Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999).
Acknowledgements
This work was supported by NIH grants CA151128 and GM118933 (R.S.); CA197996 (D.J.S.); CA120244, CA193981 and CA190533 (M.A.N.); Ruth L. Kirschstein National Research Service Award F32CA189622 (A.B.-S.); NIH grant 1F99CA21245501 (H.C.A.); Empire State Development Corporation Krabbe Disease Research Working Capital X561 and Krabbe Disease Research Capital Equipment U446 (M.L.F.); and the Jennifer Linscott Tietgen Foundation (M.A.N.). The pLV-SV4-puro lentiviral vector was obtained from P. Chumakov (Cleveland Clinic).
Author information
Authors and Affiliations
Contributions
A.B.-S., M.S.R., R.S. and M.A.N. designed the experiments and wrote the manuscript; A.B.-S. and M.S.R. performed most of the experiments and analyzed the data; C.E.F., B.C.L., L.M.P., S.M., K.M., E.E.F. and A.B. performed some of the experiments; H.C.A. performed HPLC analysis; E.H. assisted with the microscopy acquisition and analysis; D.J.S., A.V.B., E.S.K. and M.L.F. supervised part of the study; R.S. and M.A.N. conceived the initial hypothesis and supervised the study. A.B.-S. and M.S.R. contributed equally to this study. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Screening of 24 FeoB–cpYFP fusions for GTP effects on their fluorescence.
In each panel the emission spectra (500-600 nm) are plotted for the indicated fusions excited at 485nm and in the absence (blue trace) or presence (red trace) of 100μM GTP or an equal volume of GTP-free buffer (red trace). Three of the fusions (FY1+1a, FY5a+5a and FY6+6a) show large (>2-fold) reductions in emission upon binding GTP and were selected for further characterization. Fusion nomenclature is as described in main text: the number refers to the position at which the cpYFF was inserted into FeoB, with "1" corresponding to insertion after residue 35 and "6" to insertion after residue 40, and the suffix "a" indicates the addition of an "SAG" or "GT" linker at, respectively, the N- or C-terminus of the cpYFP. The last plot shows the null response of cpYFP alone to GTP.
Supplementary Figure 2 The FY5a+5a fusion is selective for GTP and dGTP.
(a) Excitation spectra (360 to 520nm; emission measured at 530nm) of FY5a+5a fusion in the absence (red trace) or presence of 4 (dark blue), 8 (yellow), 16 (green), 31 (purple), 63 (grey), 125 (pink), 250 (light blue) or 500 μM (brown) GTP. (b) As in panel (a), but using UTP rather than GTP. (c) Excitation spectra of FY5a+5a fusion in the absence of nucleotide (red), the presence of 500μM UTP (dark blue), 100μM GTP (yellow) or 100μM GTP and 500μM UTP (green) to assess the ability of UTP interfere with the GTP induced spectrum changes. (d) As in (a) but using CTP. (e) As in (c) but with CTP as interfering nucleotide. (f) As in (a) but using GMP. (g) As in (c) but with GMP as the interfering nucleotide. (h) As in (a) but using ITP. (i) As in (c) but with ITP as the interfering nucleotide. (j) As in (a) but using ATP. (k) As in (c) but with ATP as the interfering nucleotide. (l) As in (a) but using dGTP. (m) As in (a) but using GDP.
Supplementary Figure 3 The FY5a+5a fusion exhibits a robust GTP-driven ratiometric change in fluorescence across the physiologically relevant pH range.
(a-g) Plots of the fluorescence emission intensity (at 530 nm) for the FY5a+5a fusion at the indicated pH values vs. excitation wavelength in either the absence (black trace) or presence (red trace) of 1 mM GTP. The ratios of the ratiometric signals (F390ex/F484ex) +/- GTP vary from 1.7 to 5.3 across the pH 5.5 to 8.5 range.
Supplementary Figure 4 Excess GDP does not significantly interfere with GTP sensing.
Plots of 530 nM emission intensity (in AU) vs. excitation wavelength for the FY5a+5a fusion in either the absence (red trace) or presence of 0.5 mM GTP and varying concentrations of GDP as indicated (error bars show +SEM for n=5 measurements). The 485ex/400ex ratios for the data plotted are given. Addition of 0.5 mM GTP reduces the 485ex/400ex ratio by almost 3-fold while addition of 2mM GDP affects this ratio by less than 10%, and even higher GDP affect this ratio by a maximum of only 25%.
Supplementary Figure 5 Mutations in the FeoB GTP-binding site generate a set of GEVAL sensors that respond to a broad range of GTP concentrations.
Normalized F400ex/F485ex fluorescence intensity ratios (at 530 nm emission) as a function of variation in the concentration of the indicated ligands are plotted and fit to a Hill equation with offset and cooperativity parameter values of 1.0. Each data point is an average of 3-4 measurements. Error bars are ±s.e.m (bars often not visible as spread is smaller than symbol size). (a) GEVAL sensor with WT FeoB component. (b) GEVAL sensor with P12G mutation in FeoB component. (c) GEVAL sensor with P12G/S14 mutation in FeoB component. (d) GEVAL sensor with P12G/T17S mutation in FeoB component. (e) GEVAL sensor with P12G/S14A/T17S mutation in FeoB component. (f) GEVAL sensor with P12A/S14A/T17A mutation in FeoB component.
Supplementary Figure 6 Effect of pH on GEVAL fluorescence.
(a) Variation in GEVAL30 530nm fluorescence emission intensity as a function of pH upon excitation at 400nm in either the absence (black trace and square symbols) or presence (red trace and round symbols) of 0.1mM GTP. (b) Variation in GEVAL30 530nm fluorescence emission intensity as a function of pH upon excitation at 485 nm in either the absence (black trace and square symbols) or presence (red trace and round symbols) of 0.1mM GTP. (c) Variation in the ratio of GEVAL30 530nm fluorescence emission intensity as a function of pH when excited at 400 vs. 485 nm in either the absence (black trace and square symbols) or presence (red trace and round symbols) of 0.1mM GTP. In all plots the highest y-value for each data set is normalized to 1.0. Error bars show +/-SEM for n=3.
Supplementary Figure 7 GEVAL sensors exhibit identical responses to pH but varying responses to GTP depending on their GTP affinities.
(a) Emission intensity ratios for GEVAL30 at 400nm vs. 485nm excitation plotted against μM GTP at pH 5.5 (red squares), 6.0 (blue circles), 6.5 (yellow upward pointing triangles), 7.0 (green downward pointing triangles), 7.5 (purple diamonds), 8.0 (grey leftward pointing triangles), and 8.5 (pink rightward pointing triangles). (b) As in panel (a) but for GEVAL260. (c) As in (a), but for GEVAL530. (d) As in (a) but for GEVAL1150. (e) As in (a), but for GEVAL2300. (f) As in (a), but for GEVALNull. GEVALNull shows identical pH dependence as other GEVALs but no GTP sensitivity, allowing it to be used as a control to distinguish between fluorescent changes due to GTP vs. pH.
Supplementary Figure 8 Sensor affinities for GTP are relatively constant across physiologically relevant pH ranges.
(a) Keff for the GTP induced fluorescence change in GEVAL30 (red trace), GEVAL260 (blue), GEVAL530 (yellow), GEVAL1150 (green) and GEVAL2300 (purple) is plotted as a function of pH (based on data presented in Fig. 3 in main text). The measured Keff values vary by less than +/-33% across the specified pH range. Note that these values are based on experiments in which the maximum GTP concentration is 640μM, which tends to underestimate the Keff for the lower affinity sensors relative to the data (as in Table 1) that includes GTP concentrations that extend well above the sensor Keff. (b) Dynamic light scattering measurements of GEVAL30 in the absence (black trace) or presence (red trace) of 1mM GTP reveal a solution MW and narrow polydispersity profile consistent with a monodisperse monomer under both conditions.
Supplementary Figure 9 Validation of GEVAL sensors in cells.
(a) SK-Mel-103 cells were left untransduced or transduced with GEVAL530, or transfected with the pHRed sensor. Cells were imaged with regular light (phase contrast) and excited sequentially at 405nm, 458nm, 488nm, and 594nm (as described in material and methods). (b) GEVAL530-infected cells were monitored for bleaching. The same group of cells was imaged every 30sec for a total of 10 minutes. The Ex405/Ex488 ratio of 10 individual cells was calculated for each time point and the data represents the average -/+ standard deviation of these measurements. (c) SK-Mel-103 cells were transduced with two different titers for each GEVAL construct. Increasing amounts of sensors were visualized by immunoblot toward YFP and GAPDH (as a loading control). (d) cells as in (c) were treated for 48hrs with vehicle control or 1.6μM MPA. At the end of treatment cell pellets were extracted for nucleotide analysis via HPLC. The data represent the average -/+ SEM of two independent experiments. Cells as in (c) were seeded at 200K/well in 6-well plates in duplicates and treated for 48hrs with vehicle control, 0.2μM and 2μM MPA. At the end of treatment, cells were trypsinized and counted with trypan blue exclusion to determine proliferation (e) and viability (f). The data represent the average -/+ SEM of two independent experiments. (g) Cells as in (c) were imaged and the Ex405/Ex488 ratio was calculated for 30cells/population. The data represents the average -/+ SEM of 60 cells from two independent experiments. Statistics were performed by two-tailed Student t-Test, * p<0.05; n.s. not significant.
Supplementary Figure 10 Validation of pHRed sensor in cells.
(a) pH analysis by pHRed in SK-Mel-103 cells from Fig 4b,c. The approximate pH for each cell population was calculated using data provided by Dr. Gary Yellen (personal communication). The data represent the average -/+ SEM of 60 cells from two independent experiments. (b) pH analysis by pHRed in SK-Mel-103 cells from Supplementary Fig S9g. The data represent the average -/+ SEM of 60 cells from two independent experiments. (c) Cells transduced with pHRed vector were imaged sequentially at rest and after treatment with NH4Cl (20mM final) and acetic acid (10mM final) and the Ex594/Ex458 was calculated. The data represent the average -/+ SEM of 25 individual cells from two independent experiments. (d) Representative images from cells as in (c). Statistics were performed by two-tailed Student t-Test, *** p<0.0001; n.s. not significant.
Supplementary Figure 11 Time course of GEVAL sensor activity in response to IMPDH2 pharmacological inhibition by mizoribine.
(a) SK-Mel-103 cells transduced with GEVAL30, GEVAL530, or GEVALNull, and treated for 48hrs with 25μM MZR. Cells were then imaged and the Ex405/Ex488 ratio was calculated for 30cells/population. The data represent the average -/+ SEM of three independent experiments. (b) HPLC analysis of GTP in cells treated as in (a). The data represent the average -/+ SEM of 3 independent experiments. (c) SK-Mel-103 cells were transduced with GEVAL30, GEVAL530, or GEVALNull. Cells were plated on coverslip chambers and imaged (0h). Cells were then treated with 25μM MZR and cell populations were imaged at the indicated time points to calculate the Ex405/Ex488 ratio in 30 cells/population. The data represent the average -/+ SEM of 60 cells from 2 independent experiments. Quantification of Ex405/Ex488 in 10 cells/population over time for cells treated with 1.6μM MPA (d) or 25μM MZR (e). These measurements are related to the cells shown in Fig. 4. The data represent the average -/+ SEM of 20 cells from two independent experiments normalized over the GEVALNull signal and to the untreated control (0h) in each group. Statistics were performed by two-tailed Student t-Test, * p<0.05, ** p<0.001; n.s. not significant.
Supplementary Figure 12 Characteristics of the distribution of signal ratios in cells expressing GEVALs.
SK-Mel-103 cells transduced with GEVALNull, GEVAL530, or GEVAL30 were imaged and 30 cells were randomly chosen for analysis from each cell population. Pixel-by-pixel ratiometric images were created with ImageJ as described in material and methods and the signal ratio was used to calculate excess kurtosis (a) and skewness (b) for each cells. The bar and whisker plots depict median and quartiles for each parameter. Statistics were performed by two-tailed Mann-Whitney test. * p<0.05, ** p<0.001.
Supplementary Figure 13 Evaluation of GEVALs and pHRed gradients in cells.
SK-Mel-103 cells transduced with GEVALs and pHRed were imaged as described. Pixel-by-pixel ratiometric pictures for the GEVAL wavelengths (Ex450/Ex488) and the pHRed wavelengths (Ex594/Ex458) were generated in ImageJ with the Ratio Plus plug-in. pHRed ratiometric images were false-colored with the RED HOT LUT. Individual wavelengths and their ratio images are presented, along with the ratio values for 10 different areas within each cell and the corresponding approximate pH values.
Supplementary Figure 14 Evaluation of IMPDH2 dependence for shRNA effects and of GEVAL performance for HTP screenings.
(a) SK-Mel-103 cells were transduced with empty vector (pLVp) or an shRNA-resistant IMPDH2 expression vector (IMPDH2-R) and superinfected with an empty vector (pLKO) or shRNA to IMPDH2. The resulting cell populations were then transduced with the three GEVALs.IMPDH2 manipulations were verified by immunoblot and GAPDH was used to verify equal loading (b) cells as in (a) were imaged and quantified. The data represent the average -/+ SEM of two independent experiments normalized over the untreated control in each group. SK-Mel-103 cells transduced with GEVAL30 (c) or GEVAL530 (d) were seeded in a glass-bottom 96well plate (3,000cells/well; 26wells/sensor). Half of the wells were treated with vehicle control and half with 1.6μM MPA. Wells were imaged at 20X (dry lens) and the Ex405/Ex488 for 10 cells in each well was calculated; the average of the ratio of the 10 cells was used as the mean reading for that well. The Z’ factor was calculated using the formula Z'=(1-(3*(σs+σc)/|μs-μc|), where σs and σc are the standard deviations of the sample (+MPA) and the control (vehicle), respectively, and μs and μc are the means of the sample and control, respectively. Statistics were performed by two-tailed Student t-Test, * p <0.05; n.s. not significant.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Note 1.
Life Sciences Reporting Summary
Life Sciences Reporting Summary.
Rights and permissions
About this article
Cite this article
Bianchi-Smiraglia, A., Rana, M., Foley, C. et al. Internally ratiometric fluorescent sensors for evaluation of intracellular GTP levels and distribution. Nat Methods 14, 1003–1009 (2017). https://doi.org/10.1038/nmeth.4404
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4404
This article is cited by
-
GTP energy dependence of endocytosis and autophagy in the aging brain and Alzheimer’s disease
GeroScience (2023)
-
Regulation of local GTP availability controls RAC1 activity and cell invasion
Nature Communications (2021)