Abstract
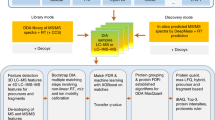
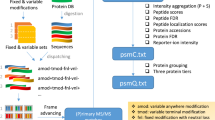
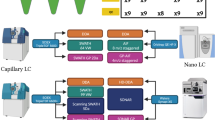
Data-independent acquisition (DIA) is an emerging mass spectrometry (MS)-based technique for unbiased and reproducible measurement of protein mixtures. DIA tandem mass spectrometry spectra are often highly multiplexed, containing product ions from multiple cofragmenting precursors. Detecting peptides directly from DIA data is therefore challenging; most DIA data analyses require spectral libraries. Here we present PECAN (http://pecan.maccosslab.org), a library-free, peptide-centric tool that robustly and accurately detects peptides directly from DIA data. PECAN reports evidence of detection based on product ion scoring, which enables detection of low-abundance analytes with poor precursor ion signal. We demonstrate the chromatographic peak picking accuracy and peptide detection capability of PECAN, and we further validate its detection with data-dependent acquisition and targeted analyses. Lastly, we used PECAN to build a plasma proteome library from DIA data and to query known sequence variants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Venable, J.D., Dong, M.-Q., Wohlschlegel, J., Dillin, A. & Yates, J.R. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 (2004).
Chapman, J.D., Goodlett, D.R. & Masselon, C.D. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom. Rev. 33, 452–470 (2014).
Röst, H.L. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 (2014).
Wang, J. et al. MSPLIT-DIA: sensitive peptide identification for data-independent acquisition. Nat. Methods 12, 1106–1108 (2015).
Ting, Y.S. et al. Peptide-centric proteome analysis: an alternative strategy for the analysis of tandem mass spectrometry data. Mol. Cell. Proteomics 14, 2301–2307 (2015).
Tsou, C.-C. et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264 (2015).
Li, Y. et al. Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files. Nat. Methods 12, 1105–1106 (2015).
Panchaud, A. et al. Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Anal. Chem. 81, 6481–6488 (2009).
Weisbrod, C.R., Eng, J.K., Hoopmann, M.R., Baker, T. & Bruce, J.E. Accurate peptide fragment mass analysis: multiplexed peptide identification and quantification. J. Proteome Res. 11, 1621–1632 (2012).
Käll, L., Canterbury, J.D., Weston, J., Noble, W.S. & MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 (2007).
Eng, J.K., Jahan, T.A. & Hoopmann, M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2012).
Gillet, L.C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012).
Beausoleil, S.A., Villén, J., Gerber, S.A., Rush, J. & Gygi, S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 (2006).
Bald, T. et al. pymzML--Python module for high-throughput bioinformatics on mass spectrometry data. Bioinformatics 28, 1052–1053 (2012).
Martens, L. et al. mzML—a community standard for mass spectrometry data. Mol. Cell. Proteomics 10, R110.000133 (2011).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Murray, K.K. et al. Definitions of terms relating to mass spectrometry (IUPAC recommendations 2013). Pure Appl. Chem. 85, 1515–1609 (2013).
Granholm, V., Navarro, J.F., Noble, W.S. & Käll, L. Determining the calibration of confidence estimation procedures for unique peptides in shotgun proteomics. J. Proteomics 80, 123–131 (2013).
Stergachis, A.B., MacLean, B., Lee, K., Stamatoyannopoulos, J.A. & MacCoss, M.J. Rapid empirical discovery of optimal peptides for targeted proteomics. Nat. Methods 8, 1041–1043 (2011).
Davis, M.T. et al. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. II. Limitations of complex mixture analyses. Proteomics 1, 108–117 (2001).
Serang, O., MacCoss, M.J. & Noble, W.S. Efficient marginalization to compute protein posterior probabilities from shotgun mass spectrometry data. J. Proteome Res. 9, 5346–5357 (2010).
Acknowledgements
The authors thank L. Käll, A.I. Nesvizhskii, N. Bandeira, and J.K. Eng for insightful discussions. This work was supported by the National Institutes of Health Grants P30 AG013280, R21 CA192983, P41 GM103533, and U54 HG008097. S.H.P. was supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, and Early Career Research Program.
Author information
Authors and Affiliations
Contributions
Y.S.T. and M.J.M. designed the experiments. Y.S.T. developed the algorithms with input from J.D.E., S.H.P., B.C.S., W.S.N., and M.J.M. Y.S.T. performed the analyses. Y.S.T. and J.G.B. acquired the data. Software was written by Y.S.T. with input from J.D.E. and B.C.S. The manuscript was written by Y.S.T. with substantial input from J.D.E., S.H.P., W.S.N., and M.J.M.
Corresponding author
Ethics declarations
Competing interests
The MacCoss Lab at the University of Washington has a sponsored research agreement with Thermo Fisher Scientific, the manufacturer of the instrumentation used in this research. Additionally, M.J.M. is a paid consultant for Thermo Fisher Scientific.
Integrated supplementary information
Supplementary Figure 1 Retention time analysis for common peptides from Comet-DDA and PECAN-DIA.
Of the 5,182 peptides commonly detected by PECAN from 4xGPF DIA data and Comet from 4xGPF DDA data, 27 peptides were identified more than 2 minutes apart.
Supplementary Figure 2 Dynamic range of DIA plasma library.
Relative concentration values of 248 plasma proteins are taken from the literature. (Source: Leigh Anderson, The Plasma Proteome Institute, Washington, DC, USA, modified from ref Mol. Cell Proteomics 1, 845–847, 2002.) Color of the dot represents the number of peptides unique to the protein or only shared by its isoforms in the DIA plasma library. Note that some literature values are measurement for protein complex or specific fragments of the protein (e.g. values for Prothrombin and Fibrinogen alpha chain), of which the intact protein concentration could be higher.
Supplementary Figure 3 Assessment of background scores estimation with 1,000 random sampling.
(a) Boxplot shows the distribution of 2,185 CVs of the RSEs from 1,000 random sampling at each decoy size. (b) The estimated background scores with 2,000 charge 2 and 2,000 charge 3 decoys for 2,185 MS/MS spectra presented over retention time. Black lines trace the median of the decoy means from 1,000 estimations by random sampling and the blue shades are segments between the 25th and 75th percentiles. (c) Bonferroni corrected p-values from Wilcoxon rank-sum tests between the 1,000 estimations using either 2,000 charge 2 or 2,000 charge 3 decoys for individual spectrum. Grey lines indicated the p-value is smaller than 0.05 and therefore rejected the null hypothesis.
Supplementary Figure 4 Evidence qualifying procedure in PECAN.
An evidence of detection (abbr. evidence) for a query peptide p at the time t is the average of the calibrated primary scores from a short period of retention time (see Methods), centered at the time t. Following this flowchart, PECAN reports a user-defined number of qualified evidence(s) that are calculated from primary scores which have never been used to calculate other qualified evidences(s).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–3 and Supplementary Notes 1–6 (PDF 4050 kb)
Reporting Summary
Life Sciences Reporting Summary (PDF 129 kb)
Rights and permissions
About this article
Cite this article
Ting, Y., Egertson, J., Bollinger, J. et al. PECAN: library-free peptide detection for data-independent acquisition tandem mass spectrometry data. Nat Methods 14, 903–908 (2017). https://doi.org/10.1038/nmeth.4390
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4390
This article is cited by
-
SeFilter-DIA: Squeeze-and-Excitation Network for Filtering High-Confidence Peptides of Data-Independent Acquisition Proteomics
Interdisciplinary Sciences: Computational Life Sciences (2024)
-
Analysis of DIA proteomics data using MSFragger-DIA and FragPipe computational platform
Nature Communications (2023)
-
A peptide-centric quantitative proteomics dataset for the phenotypic assessment of Alzheimer’s disease
Scientific Data (2023)
-
Time-resolved in vivo ubiquitinome profiling by DIA-MS reveals USP7 targets on a proteome-wide scale
Nature Communications (2021)
-
Ultra-fast proteomics with Scanning SWATH
Nature Biotechnology (2021)