Abstract
CRISPR–Cas9-induced DNA damage may have deleterious effects at high-copy-number genomic regions. Here, we use CRISPR base editors to knock out genes by changing single nucleotides to create stop codons. We show that the CRISPR-STOP method is an efficient and less deleterious alternative to wild-type Cas9 for gene-knockout studies. Early stop codons can be introduced in ∼17,000 human genes. CRISPR-STOP-mediated targeted screening demonstrates comparable efficiency to WT Cas9, which indicates the suitability of our approach for genome-wide functional screenings.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Cong, L. et al. Science 339, 819–823 (2013).
Mali, P. et al. Science 339, 823–826 (2013).
Aguirre, A.J. et al. Cancer Discov. 6, 914–929 (2016).
Hart, T. et al. Cell 163, 1515–1526 (2015).
Wang, T. et al. Science 350, 1096–1101 (2015).
Wang, T. et al. Cell 168, 890–903.e15 (2017).
Komor, A.C., Kim, Y.B., Packer, M.S., Zuris, J.A. & Liu, D.R. Nature 533, 420–424 (2016).
Chen, Z. et al. RNA 16, 1040–1052 (2010).
Shalem, O. et al. Science 343, 84–87 (2014).
Wang, T., Wei, J.J., Sabatini, D.M. & Lander, E.S. Science 343, 80–84 (2014).
Koike-Yusa, H., Li, Y., Tan, E.P., Velasco-Herrera, Mdel.C. & Yusa, K. Nat. Biotechnol. 32, 267–273 (2014).
Liao, S., Tammaro, M. & Yan, H. Nucleic Acids Res. 43, e134 (2015).
Nishida, K. & Takagi, R. Science 353, 919–921 (2016).
Kuscu, C. & Adli, M. Nat. Methods 13, 983–984 (2016).
Ma, Y. et al. Nat. Methods 13, 1029–1035 (2016).
Hess, G.T. et al. Nat. Methods 13, 1036–1042 (2016).
Kim, Y.B. et al. Nat. Biotechnol. 35, 371–376 (2017).
Kuscu, C., Tufan, T., Yang, J. & Adli, M. CRISPR-STOP: gene silencing through base editing-induced nonsense mutations. Protocol Exchange. http://dx.doi.org/10.1038/protex.2017.054 (2017).
Moreno-Mateos, M.A. et al. Nat. Methods 12, 982–988 (2015).
Singh, R., Kuscu, C., Quinlan, A., Qi, Y.J. & Adli, M. Nucleic Acids Res. 43, e118 (2015).
Kent, W.J. Genome Res. 12, 656–664 (2002).
Acknowledgements
This research was funded through a pilot project (NIDDK P50 DK096373), V Scholar award to M.A. from V Cancer Research Foundation, and UVA Cancer Center pilot project awards to M.A. We would like to thank S. Shang for his constructive feedback and critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
C.K. and M.A. designed the study; C.K. performed experiments. J.Y. and K.S. performed computational work; and T.T., M.P., X.W. and R.M. helped C.K. with experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 i-stop codon positions in the mCherry gene
The gRNAs (green) that can potentially generate stop codons from Trp (63th and 98th aa, upper panel) and Gln (47th and 114th aa, bottom panel) are shown. Potential Cytosines (or guanines) that may generate stop codons are red and corresponding inverted “Thymines” (or “Adenines”) are blue labeled.
Supplementary Figure 2 Expressing WT-Cas9 and BE3 complex in HEK293T cells
(a) Schematics of WT-Cas9 and BE3 complex expression vectors. (b) Western blot demonstrates the expression levels of WT-Cas9 and BE3 proteins after transient transfection of indicated plasmid concentrations. Protein levels were detected using the HA antibody which is fused to the C termini of both protein.
Supplementary Figure 3 BE3 activity is abolished when the catalytic activity of rat Apobec1 is double mutated
(a) Proline (P) at 29 and glutamic acid(E) at 181 were mutated into phenylalanine(F) and Glutamine (Q) respectively by using site directed mutagenesis. Sanger sequencing analyses show mutated nucleotide by red color for both loci and altered codon is underlined. (b) Mutant BE3 lost its activity for the silencing of mCherry signal in the presence of sgRNA 3 and sgRNA4 with respect to the wild type BE3. Dot plot shows flow cytometry measured percent of mCherry-negative cells upon guiding BE3(wild type) or BE3(double mutant) by sgRNA3 and 4.
Supplementary Figure 4 Effects of synonymous mutation for gene KO
(a) Sequence of the sgRNA which produces synonymous mutation in the presence of BE3 complex. There are two potential “C”s in the first eight nucleotides of protospacer. Red nucleotides show the target “C”s and blue nucleotides demonstrate the edited bases. Yellow color highlights the PAM region. (b) Control sgRNA and synonymous sgRNA are used in the presence of WT-Cas9 and BE3 complex. Dot plot shows flow cytometry measured percent of mCherry-negative cells upon guiding WT-Cas9 or BE3.Each dot represents separate experiments. P-value is calculated with unpaired t-test (***<0.001). (c)Flow cytometry analysis showing percent mCherry negative cells in low copy number cell line in the presence of WT-Cas9 and BE3. X-axis denotes mCherry signal and Y-axis demonstrates the cell number. (d) Sanger sequencing represents the nucleotide change in the Asp codon. While red codons show the wild type Asp codon in mCherry gene, blue codons represent the Asp codon after the synonymous mutation. Yellow indicates the PAM region and underlined sequence represents the sgRNA (left panel). Right panel shows sanger sequencing chromatographs of the nucleotide sequence for control and clone#6 with the edited base (*) sign shows perfect 100 % match.
Supplementary Figure 5 Sanger sequencing analysis of C to T (or G to A) conversion in Trp codon
(a) mCherry target sequence was cloned from mCherry population cells transfected with sgRNA3 and BE3. 4/9 colonies have induced stop codon (i-stop, shown by blue) at the location of Trp (63th aa) (b) Sanger DNA sequencing profiles illustrate edited nucleotides following BE3 and sgRNA3 transfection. Sanger sequencing color code is only used for Trp codon nucleotides. Arrows indicate the 4th and 8th positions of the sgRNA guiding sequence distal to PAM. (c) Sanger sequencing analysis of genetic changes induced by BE3 and WT-Cas9. mCherry target sequence was cloned from low copy mCherry cells following transfection with sgRNA4 in the presence of WT-Cas9 or BE3. 4/10 colonies have i-stop codons (shown by blue) at the location of Trp (98th aa) for BE3 transfection. There was no further C to T conversion or any other base changes in 50 bp flanking regions of sgRNA4 target site. On the other hand, WT-Cas9 introduced deletions (shown by gray dashes) in the 3/10 colonies shown in the bottom panel. sgRNA target sequences are underlined on the mCherry gene and complementary bases of PAM region on the coding strand were highlighted with yellow. Trp and i-stop codons are highlighted with red and blue respectively. Any other substitutions are labeled by brown. (*) sign shows perfect 100 % match.
Supplementary Figure 6 Identification of the copy numbers in single cell colonies from mCherry population cells
Relative mCherry copy number levels were quantified in real time-PCR by using the GAPDH primer as a control genomic region. Clone #4, #6 and #12 were used as low, medium and high mCherry copy number cell lines, respectively.
Supplementary Figure 7 Comparative analysis of KO efficiency of WT-Cas9 and BE3 mediated CRISPR-STOP approach
(a) Dot plot shows flow cytometry measured percent of mCherry-negative cells upon guiding WT-Cas9 or BE3 by four separate sgRNAs for four mCherry-targeting sgRNAs following WT-Cas9 or BE3 transfection in low and high copy number cell lines. Each dot represents seperate experiments. The black lines on each bar in the dot plots show the mean of three or more replicates. P-value is calculated with unpaired t-test (*<0.05;**<0.01). (b) Flow cytometry analysis showing percent mCherry-negative cells. X-axis denotes mCherry signal and Y-axis demonstrates the cell number. Top two rows are showing the flow analysis for mix population of mCherry cells, middle two rows show the same analysis in low copy mCherry cells and bottom two rows show the rate of silencing in high copy mCherry cells. (c) mCherry-knock out cells (yellow arrows) are shown in the merged images of fluorescence microscopy. sgRNA3 was used in the presence of BE3 in the low copy mCherry cell line.
Supplementary Figure 8 WT-Cas9 has more deleterious effects compared to the BE3 when targeted to a high copy number loci
(a)Dot plot summarizes the flow cytometry analysis of the normalized AnnexinV staining levels in low, medium and high copy number cells expressing sgRNA3 with WT-Cas9 or BE3. Each dot represents separate experiments. The black lines on each bar in the dot plots show the mean of three or more replicates. P-value is calculated with unpaired t-test (*<0.05). (b) Flow cytometry profiles are showing DAPI and AnnexinV-FITC staining. SgRNA3 and sgRNA4 were transfected to high copy mCherry cells together with WT-Cas9 or BE3. Apoptotic cells are stained by AnnexinV-FITC. Of note, cells in the center of each population, which were shown by yellow or red dots, shifted right in the WT-Cas9 treatment. Numbers in each quarter demonstrates the percent cell number. (c) Flow histogram shows AnnexinV staining levels in high copy number mCherry cells upon guiding WT-Cas9 and BE3 with control and mCherry targeting sgRNA #3.
Supplementary Figure 9 Phospho-H2Ax staining shows WT-Cas9 and BE3 induced levels of DNA damage
SgRNA3 and sgRNA4 were transfected to high copy mCherry cells together with WT-Cas9 or BE3.Scale bar represents 5 μm.
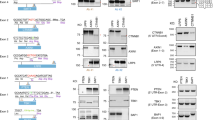
Supplementary Figure 10 CRISPR-STOP mediated gene silencing in endogenous loci.
(a, b) Green bars under exons of each gene denote the positions of sgRNAs for EHMT2 (a) and LMNB2 (b) in the upper panel. Western blot analyses of protein levels after transient transfection of the indicated sgRNAs are shown on the left sides of the bottom panel. Right sites of the bottom panel show the control protein expression of alpha-tubulin. Numbers on the left site of the images indicate the sizes of the protein marker.
Supplementary Figure 11 Comparative analysis of KO efficiency of WT-Cas9 and BE3 mediated gene silencing in endogenous LMNB2 loci
X-axis shows the BL1-mClover signal intensity and y-axis demonstrates the cell number in the flow cytometry analysis. Numbers on the bars show the percentage mClover(-) and mClover(+) cell numbers for each treatment. Top panel shows the distribution of HCT-116 control cells (no mClover). WT-Cas9 and BE3 transfected cells are shown by the middle two and bottom two panels, respectively. Numbers (#1 thru #8) show the LMNB2 sgRNA numbers.
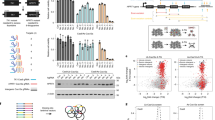
Supplementary Figure 12 Characteristics of CRISPR-STOP library
(a) Bar plots shows the total numbers of unique sgRNAs that target the four codons in the human exome. (b) The percentages of sgRNAs with different oligonucleotide stretches are shown for both the CRISPR-STOP and GeCKOv2 libraries. Only 0.9% of the sgRNAs contain 6-mer or above for i-stop, and 0.6% for GeCKOv2. (c) GC content of the sgRNAs in the CRISPR-STOP and GeCKOv2 libraries is shown. Densities of GC content between the two libraries are comparable, with the mean for i-stop slightly higher due to the specific codons targeted. (d) Nucleotide compositions at each position of the 20 base pairs of the sgRNAs were compared between the CRISPR-STOP and GeCKOv2 libraries. In general, the four nucleotides are equally distributed at each position. As expected, the distribution is skewed slightly between the 4th to 8th positions of the sgRNAs in the CRISPR-STOP library due to the specific codons targeted. (e) The schematic outline of the CRISPR-STOP mediated knockout screening with a library containing 27 sgRNAs in HEK293T cells.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 and Supplementary Tables 1–2
Supplementary Data 1
Supplementary Data 1
Supplementary Protocol
Supplementary Protocol
Rights and permissions
About this article
Cite this article
Kuscu, C., Parlak, M., Tufan, T. et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods 14, 710–712 (2017). https://doi.org/10.1038/nmeth.4327
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4327
This article is cited by
-
Escaping from CRISPR–Cas-mediated knockout: the facts, mechanisms, and applications
Cellular & Molecular Biology Letters (2024)
-
Recent advances in CRISPR-based functional genomics for the study of disease-associated genetic variants
Experimental & Molecular Medicine (2024)
-
Base editor-mediated large-scale screening of functional mutations in bacteria for industrial phenotypes
Science China Life Sciences (2024)
-
Gene-knockout by iSTOP enables rapid reproductive disease modeling and phenotyping in germ cells of the founder generation
Science China Life Sciences (2024)
-
Efficient CRISPR-Cas9 based cytosine base editors for phytopathogenic bacteria
Communications Biology (2023)