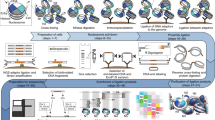
Abstract
We present Micro-C XL, an improved method for analysis of chromosome folding at mononucleosome resolution. Using long crosslinkers and isolation of insoluble chromatin, Micro-C XL increases signal-to-noise ratio. Micro-C XL maps of budding and fission yeast genomes capture both short-range chromosome fiber features such as chromosomally interacting domains and higher order features such as centromere clustering. Micro-C XL provides a single assay to interrogate chromosome folding at length scales from the nucleosome to the full genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Dekker, J., Marti-Renom, M.A. & Mirny, L.A. Nat. Rev. Genet. 14, 390–403 (2013).
Dekker, J. & Misteli, T. Cold Spring Harb. Perspect. Biol. 7, a019356 (2015).
Horn, P.J. & Peterson, C.L. Science 297, 1824–1827 (2002).
Friedman, N. & Rando, O.J. Genome Res. 25, 1482–1490 (2015).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Science 295, 1306–1311 (2002).
Lieberman-Aiden, E. et al. Science 326, 289–293 (2009).
Denker, A. & de Laat, W. Genes Dev. 30, 1357–1382 (2016).
Lajoie, B.R., Dekker, J. & Kaplan, N. Methods 72, 65–75 (2015).
Hsieh, T.H. et al. Cell 162, 108–119 (2015).
Duan, Z. et al. Nature 465, 363–367 (2010).
Scherrer, R., Louden, L. & Gerhardt, P. J. Bacteriol. 118, 534–540 (1974).
Marbouty, M. et al. eLife 3, e03318 (2014).
Mizuguchi, T. et al. Nature 516, 432–435 (2014).
Henikoff, S., Henikoff, J.G., Sakai, A., Loeb, G.B. & Ahmad, K. Genome Res. 19, 460–469 (2009).
Nagano, T. et al. Genome Biol. 16, 175 (2015).
Gavrilov, A.A. et al. Nucleic Acids Res. 41, 3563–3575 (2013).
Rao, S.S. et al. Cell 159, 1665–1680 (2014).
Ansari, A. & Hampsey, M. Genes Dev. 19, 2969–2978 (2005).
O'Sullivan, J.M. et al. Nat. Genet. 36, 1014–1018 (2004).
Lee, K., Hsiung, C.C., Huang, P., Raj, A. & Blobel, G.A. Genes Dev. 29, 1992–1997 (2015).
Tsankov, A.M., Thompson, D.A., Socha, A., Regev, A. & Rando, O.J. PLoS Biol. 8, e1000414 (2010).
Yuan, G.-C. et al. Science 309, 626–630 (2005).
Rando, O., Hsieh, T.-H., Fundenberg, G. & Goloborodko, A. Protocol Exchange http://dx.doi.org/10.1038/protex.2016.058 (2016).
Imakaev, M. et al. Nat. Methods 9, 999–1003 (2012).
Naumova, N. et al. Science 342, 948–953 (2013).
Imakaev, M.V., Fudenberg, G. & Mirny, L.A. FEBS Lett. 589, 3031–3036 (2015).
Acknowledgements
We thank L. Mirny and members of the Rando lab for insightful discussions. Work was supported in part by NIH grant GM079205 to O.J.R. G.F. and A.G. were supported by 4D Nucleome Program grants R01 GM114190 and U54 DK107980. T.-H.S.H. is an HHMI international student research fellow.
Author information
Authors and Affiliations
Contributions
T.-H.S.H. and O.J.R. conceived the study. T.-H.S.H. performed all experiments. G.F. and T.-H.S.H. analyzed data. T.-H.S.H., G.F., A.G., and O.J.R. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–18 and Supplementary Protocol (PDF 10017 kb)
Supplementary Data
Read numbers and interactions vs. distance for all datasets. (XLSX 813 kb)
Rights and permissions
About this article
Cite this article
Hsieh, TH., Fudenberg, G., Goloborodko, A. et al. Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome. Nat Methods 13, 1009–1011 (2016). https://doi.org/10.1038/nmeth.4025
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4025
This article is cited by
-
In vitro reconstitution of chromatin domains shows a role for nucleosome positioning in 3D genome organization
Nature Genetics (2024)
-
Mapping nucleosome-resolution chromatin organization and enhancer-promoter loops in plants using Micro-C-XL
Nature Communications (2024)
-
Ultrastructure and fractal property of chromosomes in close-to-native yeast nuclei visualized using X-ray laser diffraction
Scientific Reports (2023)
-
Visualization and data exploration of chromosome conformation capture data using Voronoi diagrams with v3c-viz
Scientific Reports (2023)
-
RNA polymerase II dynamics shape enhancer–promoter interactions
Nature Genetics (2023)