Abstract
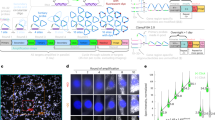
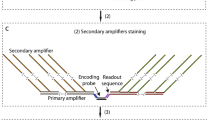
Sequential barcoded fluorescent in situ hybridization (seqFISH) allows large numbers of molecular species to be accurately detected in single cells, but multiplexing is limited by the density of barcoded objects. We present correlation FISH (corrFISH), a method to resolve dense temporal barcodes in sequential hybridization experiments. Using corrFISH, we quantified highly expressed ribosomal protein genes in single cultured cells and mouse thymus sections, revealing cell-type-specific gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lubeck, E., Coskun, A.F., Zhiyentayev, T., Ahmad, M. & Cai, L. Nat. Methods 11, 360–361 (2014).
Chen, K.H., Boettiger, A.N., Moffitt, J.R., Wang, S. & Zhuang, X. Science 348, aaa6090 (2015).
Lee, J.H. et al. Science 343, 1360–1363 (2014).
Ke, R. et al. Nat. Methods 10, 857–860 (2013).
Lubeck, E. & Cai, L. Nat. Methods 9, 743–748 (2012).
Holden, S.J., Uphoff, S. & Kapanidis, A.N. Nat. Methods 8, 279–280 (2011).
Zhu, L., Zhang, W., Elnatan, D. & Huang, B. Nat. Methods 9, 721–723 (2012).
Schwille, P. Cell Biochem. Biophys. 34, 383–408 (2001).
Kettling, U., Koltermann, A., Schwille, P. & Eigen, M. Proc. Natl. Acad. Sci. USA 95, 1416–1420 (1998).
Costantino, S., Comeau, J.W.D., Kolin, D.L. & Wiseman, P.W. Biophys. J. 89, 1251–1260 (2005).
Kondrashov, N. et al. Cell 145, 383–397 (2011).
Raj, A., van den Bogaard, P., Rifkin, S.A., van Oudenaarden, A. & Tyagi, S. Nat. Methods 5, 877–879 (2008).
Signer, R.A., Magee, J.A., Salic, A. & Morrison, S.J. Nature 509, 49–54 (2014).
Ito, Y. et al. Science 346, 363–368 (2014).
Xue, S. & Barna, M. Nat. Rev. Mol. Cell Biol. 13, 355–369 (2012).
Magde, D., Elson, E. & Webb, W.W. Phys. Rev. Lett. 29, 705–708 (1972).
Gerdes, M.J. et al. Proc. Natl. Acad. Sci. USA 110, 11982–11987 (2013).
Murray, E. et al. Cell 163, 1500–1514 (2015).
Gong, H. et al. Bioconjug. Chem. 27, 217–225 (2016).
Fan, R. et al. Nat. Biotechnol. 26, 1373–1378 (2008).
Xue, M. et al. J. Am. Chem. Soc. 12, 4066–4069 (2015).
Acknowledgements
We thank J. Linton from the Elowitz laboratory (Caltech) for providing cell lines and M. Yui from the Rothenberg Laboratory (Caltech) for the intact thymus organ. We appreciate the help of the City of Hope Pathology Core to slice thymus into sections. This work is funded by US National Institute of Health single-cell analysis program award R01HD075605. A.F.C. is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Author information
Authors and Affiliations
Contributions
A.F.C. and L.C. designed the project and wrote the manuscript. L.C. supervised the project.
Corresponding author
Ethics declarations
Competing interests
L.C. and A.F.C. declare conflict of interests and have filed a patent application.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1-22 and Supplementary Note (PDF 4857 kb)
Supplementary Software
Correlation FISH software package v1.0 (ZIP 16177 kb)
Rights and permissions
About this article
Cite this article
Coskun, A., Cai, L. Dense transcript profiling in single cells by image correlation decoding. Nat Methods 13, 657–660 (2016). https://doi.org/10.1038/nmeth.3895
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3895
This article is cited by
-
Visualization and modeling of inhibition of IL-1β and TNF-α mRNA transcription at the single-cell level
Scientific Reports (2021)
-
Multiplex bioimaging of single-cell spatial profiles for precision cancer diagnostics and therapeutics
npj Precision Oncology (2020)
-
The diversity of GABAergic neurons and neural communication elements
Nature Reviews Neuroscience (2019)
-
Digital posters for interactive cellular media and bioengineering education
Communications Biology (2019)
-
Enhanced mRNA FISH with compact quantum dots
Nature Communications (2018)