Abstract
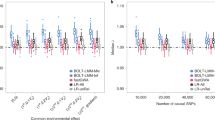
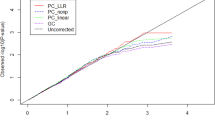
In epigenome-wide association studies (EWAS), different methylation profiles of distinct cell types may lead to false discoveries. We introduce ReFACTor, a method based on principal component analysis (PCA) and designed for the correction of cell type heterogeneity in EWAS. ReFACTor does not require knowledge of cell counts, and it provides improved estimates of cell type composition, resulting in improved power and control for false positives in EWAS. Corresponding software is available at http://www.cs.tau.ac.il/~heran/cozygene/software/refactor.html.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jaffe, A.E. & Irizarry, R.A. Genome Biol. 15, R31 (2014).
Zou, J., Lippert, C., Heckerman, D., Aryee, M. & Listgarten, J. Nat. Methods 11, 309–311 (2014).
Houseman, E.A. et al. BMC Bioinformatics 13, 86 (2012).
Reinius, L.E. et al. PLoS One 7, e41361 (2012).
Houseman, E.A., Molitor, J. & Marsit, C.J. Bioinformatics 30, 1431–1439 (2014).
Koestler, D.C. et al. Epigenetics 8, 816–826 (2013).
Pino-Yanes, M. et al. J. Allergy Clin. Immunol. 135, 228–235 (2015).
Liu, Y. et al. Nat. Biotechnol. 31, 142–147 (2013).
Goronzy, J.J. et al. J. Clin. Invest. 94, 2068–2076 (1994).
Horvath, S. Genome Biol. 14, R115 (2013).
Singmann, P. et al. Epigenetics Chromatin 8, 43 (2015).
Zeilinger, S. et al. PLoS One 8, e63812 (2013).
Shoemaker, R., Deng, J., Wang, W. & Zhang, K. Genome Res. 20, 883–889 (2010).
Wagner, J.R. et al. Genome Biol. 15, R37 (2014).
Halko, N., Martinsson, P.G. & Tropp, J.A. SIAM Rev. 53, 217–288 (2011).
Abraham, G. & Inouye, M. PLoS One 9, e93766 (2014).
Maksimovic, J., Gordon, L. & Oshlack, A. Genome Biol. 13, R44 (2012).
Johnson, W.E., Li, C. & Rabinovic, A. Biostatistics 8, 118–127 (2007).
Aryee, M.J. et al. Bioinformatics 30, 1363–1369 (2014).
Acknowledgements
The authors acknowledge the families and patients for their participation and thank the numerous health care providers and community clinics involved for their support and participation in GALA II. The research was partially supported by the Edmond J. Safra Center for Bioinformatics at Tel-Aviv University. E.H. and E.R. were supported in part by the Israel Science Foundation (Grant 1425/13), Y.B. and E.H. by the United States-Israel Binational Science Foundation (Grant 2012304). Y.B., E.H., and E.R. were partially supported by the German-Israeli Foundation (Grant 1094-33.2/2010) and by the National Science Foundation (Grant III-1217615). E.R. was supported by Len Blavatnik and the Blavatnik Family Foundation. E.E. was supported by National Science Foundation grants 1065276, 1302448, 1320589 and 1331176, and National Institutes of Health grants R01-GM083198, R01-ES021801, R01-MH101782, R01-ES022282 and U54EB020403. This research was supported in part by the Sandler Foundation, the American Asthma Foundation, and the National Institutes of Health (R01 ES015794, R01 HL088133, M01 RR000083, R01 HL078885, R01 HL104608, P60 MD006902, U19 AI077439, M01 RR00188). N.Z. was supported in part by an NIH career development award from the NHLBI (K25HL121295). J.G. was supported in part by NIH training grants GM007546, K23HL111636, and KL2TR000143 and by the Hewett Fellowship.
Author information
Authors and Affiliations
Contributions
E.R. and E.H. designed research, performed research, contributed analytic tools, analyzed data and wrote the paper. N.Z. and E.E. helped with experimental design, data interpretation, and drafting of the paper. Y.B. and J.Z. contributed expertise. C.E., D.H., J.G., S.O. and E.G.B. generated and contributed the data. D.H. also performed quality control analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16, Supplementary Tables 1–4 and Supplementary Note 1 (PDF 1529 kb)
Supplementary Text
NMETH-BC25074F.pdf (PDF 607 kb)
Rights and permissions
About this article
Cite this article
Rahmani, E., Zaitlen, N., Baran, Y. et al. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods 13, 443–445 (2016). https://doi.org/10.1038/nmeth.3809
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3809
This article is cited by
-
An overview of DNA methylation-derived trait score methods and applications
Genome Biology (2023)
-
A variability in response of osteoclasts to zoledronic acid is mediated by smoking-associated modification in the DNA methylome
Clinical Epigenetics (2023)
-
Epigenomic response to albuterol treatment in asthma-relevant airway epithelial cells
Clinical Epigenetics (2023)
-
Identification of key DNA methylation changes on fasting plasma glucose: a genome-wide DNA methylation analysis in Chinese monozygotic twins
Diabetology & Metabolic Syndrome (2023)
-
A novel approach to risk exposure and epigenetics—the use of multidimensional context to gain insights into the early origins of cardiometabolic and neurocognitive health
BMC Medicine (2023)