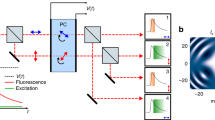
Abstract
We introduce a pattern-matching technique for efficient identification of fluorophore ratios in complex multidimensional fluorescence signals using reference fluorescence decay and spectral signature patterns of individual fluorescent probes. Alternating pulsed laser excitation at three different wavelengths and time-resolved detection on 32 spectrally separated detection channels ensures efficient excitation of fluorophores and a maximum gain of fluorescence information. Using spectrally resolved fluorescence lifetime imaging microscopy (sFLIM), we were able to visualize up to nine different target molecules simultaneously in mouse C2C12 cells. By exploiting the sensitivity of fluorescence emission spectra and the lifetime of organic fluorophores on environmental factors, we carried out fluorescence imaging of three different target molecules in human U2OS cells with the same fluorophore. Our results demonstrate that sFLIM can be used for super-resolution multi-target imaging by stimulated emission depletion (STED).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lichtman, J.W. & Conchello, J.A. Fluorescence microscopy. Nat. Methods 2, 910–919 (2005).
Minsky, M. Microscopy apparatus. US patent 3,013,467 (1961).
Pawley, J.B. (ed.) Handbook of Biological Confocal Microscopy 3rd edn. (Springer, 2006).
Denk, W., Strickler, J.H. & Webb, W.W. Two-photon laser scanning microscopy. Science 248, 73–76 (1990).
Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998).
Shaner, N.C., Steinbach, P.A. & Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909 (2005).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003).
Miller, L.W. & Cornish, V.W. Selective chemical labeling of proteins in living cells. Curr. Opin. Chem. Biol. 9, 56–61 (2005).
Johnson, I. & Spence, M.T.Z. (eds.) The Molecular Probes Handbook: A Guide to Fluorescent Probes and Labeling Technologies 11th edn., (2010).
Moerner, W.E. & Orrit, M. Illuminating single molecules in condensed matter. Science 283, 1670–1676 (1999).
Weiss, S. Fluorescence spectroscopy of single biomolecules. Science 283, 1676–1683 (1999).
Jameson, D.M. & Ross, J.A. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 110, 2685–2708 (2010).
Liyanage, M. et al. Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 14, 312–315 (1996).
Garini, Y., Gil, A., Bar-Am, I., Cabib, D. & Katzir, N. Signal to noise analysis of multiple color fluorescence imaging microscopy. Cytometry 35, 214–226 (1999).
Tsurui, H. et al. Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. J. Histochem. Cytochem. 48, 653–662 (2000).
Bastiaens, P.I. & Squire, A. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9, 48–52 (1999).
Chang, C.W., Sud, D. & Mycek, M.A. Fluorescence lifetime imaging microscopy. Methods Cell Biol. 81, 495–524 (2007).
Elangovan, M., Day, R.N. & Periasami, A. Nanosecond fluorescence resonance energy transfer—fluorescence lifetime imaging microscopy to localize the protein interactions in a single living cell. J. Microsc. 205, 3–14 (2002).
Wahl, M., Koberling, F., Patting, M., Rahn, H. & Erdmann, R. Time-resolved confocal fluorescence imaging and spectroscopy system with single molecule sensitivity and sub-micrometer resolution. Curr. Pharm. Biotechnol. 5, 299–308 (2004).
Gregor, I. & Patting, M. Pattern-based linear unmixing for efficient and reliable analysis of multicomponent TCSPC data. In Advanced Photon Counting: Applications, Methods, Instrumentation (eds. Kapusta, P., Wahl, M. & Erdmann, R.) 241–263 (Springer, 2015).
Tinnefeld, P., Herten, D.P. & Sauer, M. Photophysical dynamics of single dye molecules studied by spectrally-resolved fluorescence lifetime imaging microscopy (SFLIM). J. Phys. Chem. A 105, 7989–8003 (2001).
Knemeyer, J.P., Herten, D.P. & Sauer, M. Detection and identification of single molecules in living cells using spectrally-resolved fluorescence lifetime imaging microscopy (SFLIM). Anal. Chem. 75, 2147–2153 (2003).
Heilemann, M. et al. Multistep energy transfer in single molecular photonic wires. J. Am. Chem. Soc. 126, 6514–6515 (2004).
Becker, W. et al. Fluorescence lifetime images and correlation spectra obtained by multidimensional time-correlated single photon counting. Microsc. Res. Tech. 69, 186–195 (2006).
Fereidouni, F., Reitsma, K. & Gerritsen, H.C. High speed multispectral fluorescence lifetime imaging. Opt. Express 21, 11769–11782 (2013).
Enderlein, J. et al. A maximum likelihood estimator to distinguish single molecules by their fluorescence decays. Chem. Phys. Lett. 270, 464–470 (1997).
Lieberwirth, U. et al. Multiplex dye DNA sequencing in capillary gel electrophoresis by diode laser-based time-resolved fluorescence detection. Anal. Chem. 70, 4771–4779 (1998).
Enderlein, J. & Sauer, M. Optimal algorithm for single-molecule identification with time-correlated single-photon counting. J. Phys. Chem. A 105, 48–53 (2001).
Hanley, Q.S. Spectrally-resolved fluorescent lifetime imaging. J. R. Soc. Interface 6 (suppl. 1), S83–S92 (2009).
Zhou, Y., Dickenson, J.M. & Hanley, Q.S. Imaging lifetime and anisotropy spectra in the frequency domain. J. Microsc. 234, 80–88 (2009).
Fereidouni, F., Bader, A.N. & Gerritsen, H.C. Spectral phasor analysis allows rapid and reliable unmixing of fluorescence microscopy spectral images. Opt. Express 20, 12729 (2012).
Klar, T.A., Jakobs, S., Dyba, M., Egner, A. & Hell, S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 97, 8206–8210 (2000).
Knemeyer, J.P., Marmé, N. & Sauer, M. Probes for detection of specific DNA sequences at the single molecule level. Anal. Chem. 72, 3717–3724 (2000).
Heinlein, T., Knemeyer, J.P., Piestert, O. & Sauer, M. Nucleobase-specific quenching of fluorescent dyes in DNA-hairpins. J. Phys. Chem. B 107, 7957–7964 (2003).
Marmé, N., Knemeyer, J.P., Wolfrum, J. & Sauer, M. Inter- and intramolecular fluorescence quenching of organic dyes by tryptophan. Bioconjug. Chem. 14, 1133–1139 (2003).
Doose, S., Neuweiler, H. & Sauer, M. A close look at fluorescence quenching of organic dyes by tryptophan. ChemPhysChem 6, 2277–2285 (2005).
Donnert, G. et al. Two-color far-field fluorescence nanoscopy. Biophys. J. 92, L67–L69 (2007).
Meyer, L. et al. Dual-color STED microscopy at 30-nm focal-plane resolution. Small 4, 1095–1100 (2008).
Schmitt, F.-J. et al. eGFP-pHsens as a highly sensitive fluorophore for cellular pH determination by fluorescence lifetime imaging microscopy (FLIM). Biochim. Biophys. Acta 1837, 1581–1593 (2014).
Alsheimer, M., von Glasenapp, E., Schnölzer, M., Heid, H. & Benavente, R. Meiotic lamin C2: the unique amino-terminal hexapeptide GNAEGR is essential for nuclear envelope association. Proc. Natl. Acad. Sci. USA 97, 13120–13125 (2000).
Löschberger, A., Niehörster, T. & Sauer, M. Click chemistry for the conservation of cellular structures and fluorescent proteins: ClickOx. Biotechnol. J. 9, 693–697 (2014).
Kullback, S. & Leibler, R.A. On information and sufficiency. Ann. Stat. 22, 79–86 (1951).
Lee, D.D. & Seung, H.S. Algorithms for non-negative matrix factorization. In Advances in Neural Information Processing Systems 13 (eds. Leen, T.K., Dietterich, T.G. & Tresp, V.) 556–562 (MIT Press, 2000).
Neher, R.A. et al. Blind source separation techniques for the decomposition of multiply labeled fluorescence images. Biophys. J. 96, 3791–3800 (2009).
Acknowledgements
We thank T. Krüger and M. Alsheimer (Department of Cell and Developmental Biology, Biozentrum Universität Würzburg, Würzburg, Germany) for providing plasmids and antibodies. This work was supported by the Biophotonics Initiative of the German Bundesministerium für Bildung und Forschung (BMBF grants 13N10432 and 13N12781 to T.N., A.L. and M.S.).
Author information
Authors and Affiliations
Contributions
T.N., A.L., I.G., M.S. and B.K. designed experiments. T.N., A.L., I.G., B.K., H.-J.R., M.P. and F.K. generated and processed data. T.N., A.L., J.E., I.G., B.K. and M.S. wrote, edited and approved the final draft of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Five cellular structures of U2OS cells labeled with five fluorophores excitable at 532 nm.
(a) Mere intensity image. (b) sFLIM composite image consisting of (c) F-actin stained with ATTO 520 phalloidin (green), (d) transfection of fibrillarin-mRFP mainly localizing in the nucleoli (blue), (e) microtubules stained with Alexa Fluor 555-labeled rabbit anti-β-tubulin direct antibody (magenta), (f) Golgi stained with primary rabbit antibody against giantin and Alexa Fluor 532-labeled goat anti-rabbit secondary antibody (cyan), and (g) mitochondria stained with MitoTracker Orange (yellow). (h,i) The five corresponding reference patterns: (h) fluorescence decays after the three laser pulses and (i) emission spectra with some gaps where the excitation light is filtered out. The brightest pixels belong to 7,181 (a) / 6,798 / 6,308 / 3,514 / 4,904 / 2,967 (c–g) photons. Scale bars, 10 µm.
Supplementary Figure 2 Five cellular structures of U2OS cells labeled with five fluorophores excitable at 640 nm.
(a) Mere intensity image. (b) sFLIM composite image consisting of (c) F-actin stained with ATTO 655 phalloidin (green), (d) nucleus stained with DNA intercalating DRAQ5 (blue), (e) nucleoli stained with primary mouse antibody against fibrillarin and Alexa Fluor 633-labeled goat anti-mouse secondary antibody (white), (f) Golgi stained with primary rabbit antibody against giantin and Alexa Fluor 700-labeled goat anti-rabbit secondary antibody (yellow), and (g) microtubules stained with Alexa Fluor 647-labeled rabbit anti-α-tubulin direct antibody (magenta). (h,i) The five corresponding reference patterns: (h) fluorescence decays after the three laser pulses and (i) emission spectra. The brightest pixels belong to 4,139 (a) / 1,895 / 2,159 / 942 / 4,129 / 1,509 (c–g) photons. Scale bars, 10 µm.
Supplementary Figure 3 Using less information for sFLIM analysis.
U2OS cell labeled with five fluorophores excitable at 640 nm (Supplementary Fig. 2). (a) sFLIM image using the total information available (spectral and lifetime information). (b) Fluorescence induced by 485 nm and 532 nm lasers was ignored in data analysis and only the fluorescence from the most appropriate laser at 640 nm was used. (c) Only the spectral information (i.e. spectral un-mixing), and (d) only the lifetime information upon irradiation at 640 nm was used (i.e. FLIM). Scale bars, 10 µm.
Supplementary Figure 4 Three different structures in U2OS cells labeled with Alexa Fluor 647.
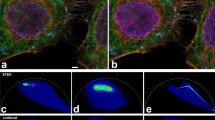
The three different staining strategies cause small changes in the dye's fluorescence properties, i.e., lifetime (f) and spectrum (g). These changes are sufficient to clearly distinguish between all three structures: (c) Golgi stained with primary rabbit antibody against giantin and Alexa Fluor 647-labeled goat anti-rabbit secondary antibody (magenta), (d) F‑actin stained with Alexa Fluor 647 phalloidin (green), and (e) EdU (nascent DNA) inside the nucleus stained via click chemistry with Alexa Fluor 647 azide (blue). (b) shows the resulting sFLIM image and (a) is the mere intensity image. The brightest pixels belong to 3,396 (a) / 3,396 / 1,204 / 2,553 (c–e) photons. Scale bars, 10 µm.
Supplementary Figure 5 Dealing with nonspecific staining.
U2OS cells were labeled with ATTO 488 phalloidin (green), Alexa Fluor 488 indirect immunofluorescence against giantin (magenta), and EdU (nascent DNA) clicked to ATTO 488 azide (blue). (a) Resulting sFLIM image. Here not only the nucleus is blue as intended, but there are also some vesicle-like spots clearly visible in the cytoplasm. They most likely consist of free ATTO 488 azide. After introduction of a new pattern for this unspecific staining, the yellow “bubbles” are clearly separated from the blue nucleus (b). Optionally one could remove the unspecific signal. Scale bars, 10 µm.
Supplementary Figure 6 Single staining of U2OS cells with MitoTracker Orange.
(a) Overall fluorescence intensity image. Using sFLIM three slightly different reference patterns (f,g) can be isolated, and three different cellular structures visualized: (c) mitochondria (magenta), (d) endosomes (green), and (e) unspecific staining, accumulated inside the nucleoli (yellow). (b) sFLIM composite image. The brightest pixels belong to 3,718 (a) / 3,275 / 2,496 / 1,223 (c–e) photons. Scale bars, 10 µm.
Supplementary Figure 7 Testing the accuracy of the pattern-matching algorithm.
Fluorescence decay patterns of three different secondary antibodies labeled with Cy3, TRITC, and TexasRed, respectively. The decays are not mono-exponential and show average lifetimes of 1.12 ns, 2.30 ns, and 3.36 ns. These decay patterns (without spectral information) were used to simulate signals of varying contributions of the patterns.
Supplementary Figure 8 Confidence levels of the algorithm.
Error of the mean amplitude (a,c,e) and the standard deviation of the amplitude (b,d,f) of Cy3 (a,b), TRITC (c,d) and TexasRed (e,f) in ternary mixtures of the three dyes. The photon numbers in the simulations are 200, 400, 800, 1,600, 2,400, and 3,600 (starting from bottom and going counter-clockwise). The color bar shows the respective values as percentage of the total number of photons per pixel.
Supplementary Figure 9 Confidence levels of the algorithm in comparison to a brute-force minimum search of the maximum-likelihood value.
Error of the mean amplitude (a,c) and the standard deviation of the amplitude (b,d) of TexasRed in ternary mixtures of dyes. (a,b) show the results of our algorithm (see Suppl. Figs. 7e,f), while (c,d) result from a brute force minimum search of the maximum likelihood value. The photon numbers in the simulations are 200, 400, 800, 1,600, 2,400, and 3,600 (starting from bottom and going counter-clockwise). The color bar shows the respective values as percentage of the total number of photons per pixel.
Supplementary Figure 10 Appropriate average patterns.
We created different reference patterns from singly- and multi-stained samples to analyze Golgi, nascent DNA and F-actin, all labeled with ATTO 488 (Fig. 4). We then calculated average patterns and standard deviations of all patterns that we classified to be appropriate, i.e. that comply with the requirements described in the Online Methods. Black lines represent average patterns for giantin/Golgi (a), phalloidin/F-actin (b), and EdU/DNA (c). Grey areas illustrate the average patterns plus and minus their standard deviation. 4/6/3 patterns have been used (a–c). Colored lines show examples of inappropriate patterns.
Supplementary Figure 11 Comparison of the results of the algorithm for different sets of three patterns.
Scatter plots showing the correlation of the retrieved amplitudes of the respective component Golgi/giantin (left), F-actin/phalloidin (middle), and DNA/EdU (right). The reference amplitude was obtained using the mean patterns for all three components as shown in Supplementary Fig. 9. The legends indicate the patterns that were altered by plus/minus standard deviation (Supplementary Fig. 9) together with the equation of the linear model describing the data. Correlation coefficients of each data-set with the linear model were larger than 0.99. The dotted lines indicate the shot noise limits A ± square root (A).
The top row (a–c) shows the effect if instead of the reference pattern the pattern plus or minus the standard deviation is used. The effect is strongest for the amplitude of the DNA stain. The linear regression shows a systematic decrease of the amplitude by 6% and 7%, respectively. For the other components, actin and giantin, the effect is in the order of up to 3%. It is usually found, that the pattern showing an average lifetime between the values of the other two components is most sensitive to deviations of the pattern. The middle row (d–f) shows how the amplitude of the respective component responds to changes of the other patterns. The deviations are all in the order of 1% to 3% and even if the two other patterns (bottom row, g–i) are altered, the values do not increase. In conclusion, we see the most pronounced effect, if the pattern of the component deviates from its optimal shape. However, if the deviations are within the standard deviation of the reference pattern, the observed effects are on average in the order of 5%. Furthermore, the analysis shows that the method is non-problematic and shows linear behavior within the scope of this analysis.
Supplementary Figure 12 Comparison of results when two independently acquired sets of reference patterns are used.
(a,b) Reference patterns, in magenta / green / blue as used in Fig. 4 (3x ATTO 488), and in brown / dark green / dark blue a second set of patterns, generated from one singly-stained sample (giantin/Golgi) and from singly-stained parts of two multi-stained images (phalloidin/F-actin and EdU/DNA). (c–f) Results obtained for the second set of pattern (Golgi, F-actin, DNA, Composite), and (g–i) pixel intensity differences in comparison to Fig. 4 (Golgi, F-actin, DNA). Here green indicates an increase and magenta a decrease in signal intensity. Numbers in brackets represent the photon counts of the brightest pixels. The mean of the absolute value of the difference is 15.0, 26.7, and 15.8 photons per pixel (g–i). (j) Mere intensity image. Scale bars, 10 µm.
Supplementary Figure 13 Comparison of results obtained from reference patterns of bright and dim regions.
(a,b) Reference patterns, in magenta / green / blue as used in Fig. 4 (3x ATTO 488), and in cyan and dark green different patterns for phalloidin/F-actin generated from very bright and very dim pixels, respectively. The pixel selection is depicted in (k) and (s) as red parts of the images. (c) Corresponding intensity image of the 3x ATTO 488 sample used for pattern extraction. Patterns for giantin/Golgi and EdU/DNA were the same as for Fig. 4. (d–g) Resulting image obtained using a bright phalloidin/F-actin pattern (Golgi, F-actin, DNA, composite). (h–j) Difference image to Fig. 4 (Golgi, F-actin, DNA), where green indicates an increased signal and magenta a decreased signal. Numbers in brackets represent the photon counts of the brightest pixels. The mean of the absolute value of the difference is 9.4, 21.9, and 6.1 photons per pixel (h–j). (l–o) Images using a dim phalloidin/F-actin pattern (Golgi, F-actin, DNA, composite). (p–r) Difference image to Fig. 4 (Golgi, F-actin, DNA). The mean of the absolute value of the difference is 20.4, 33.2, and 13.1 photons per pixel (p–r). Scale bars, 10 µm.
Supplementary Figure 14 Influence of inappropriate reference patterns.
(a,b) Reference patterns, in magenta / green / blue as used in Fig. 4 (3x ATTO 488), and in dark green and cyan inappropriate patterns generated for phalloidin/F-actin and EdU/DNA, respectively, from an older sample. The two patterns do not comply with all the requirements described in the Online Methods, i.e. the samples were prepared some days earlier than the multi-stained sample, possibly resulting in different staining conditions and sample decomposition. In addition they were measured one day after the multi-stained sample, possibly under slightly different optical and electronical settings. (c–f) Composite image (Golgi, F-actin, DNA) using an improper pattern for phalloidin/F-actin together with suited patterns for giantin/Golgi and EdU/DNA as used in Fig. 4. (g–i) Difference image to Fig. 4 (Golgi, F-actin, DNA), where green indicates an increase and magenta a decrease in signal. Numbers in brackets represent the photon counts of the brightest pixels. The mean of the absolute value of the difference is 125.6, 131.1, and 46.3 photons per pixel (g–i). (j–m) Composite image (Golgi, F-actin, DNA) using an improper pattern for EdU/DNA together with suited patterns for giantin/Golgi and phalloidin/F-actin as used in Fig. 4. (n–p) Difference image to Fig. 4 (Golgi, F-actin, DNA). The mean of the absolute value of the difference is 18.7, 134.5, and 120.5 photons per pixel (n–p). (q) Mere intensity image. Scale bars, 10 µm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Tables 1 and 2, and Supplementary Notes 1–3 (PDF 3860 kb)
Supplementary Software
sFLIM software (ZIP 116 kb)
Rights and permissions
About this article
Cite this article
Niehörster, T., Löschberger, A., Gregor, I. et al. Multi-target spectrally resolved fluorescence lifetime imaging microscopy. Nat Methods 13, 257–262 (2016). https://doi.org/10.1038/nmeth.3740
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3740
This article is cited by
-
Multicolor lifetime imaging and its application to HIV-1 uptake
Nature Communications (2023)
-
Image segmentation and separation of spectrally similar dyes in fluorescence microscopy by dynamic mode decomposition of photobleaching kinetics
BMC Bioinformatics (2022)
-
Extra kinetic dimensions for label discrimination
Nature Communications (2022)
-
Generative adversarial network enables rapid and robust fluorescence lifetime image analysis in live cells
Communications Biology (2022)
-
Fluorescence lifetime DNA-PAINT for multiplexed super-resolution imaging of cells
Communications Biology (2022)